- Volume 61 , Number 3
- Page: 389–93
Detection of Mycobacterium leprae by the polymerase chain reaction in nasal swabs of leprosy patients and their contacts
ABSTRACT
Nose swabs f rom 4 paucibacillary (PB) and 8 multibacillary (MB) leprosy patients and their contacts were tested for the presence oí Mycobacterium leprae by two polymerase chain reactions (PCR); 30% of the samples contained inhibitors for the PCR, 1 of 52 (1.9%) swabs and 13 of 164 (7.9%) swabs were positive for M. leprae among contacts of PB and MB patients, respectively. Since this difference is not significant, and some positives were found among contacts of MB patients treated and cured of their infection, it is concluded that the observed infections are community acquired.RÉSUMÉ
Les frottis des fosses nasales de 4 patients lépreux paucibacillaires (PB) et de 8 multibacillaires (MB) et de leurs contacts ont été testés pour la présence de Mycobacterium leprae par deux réactions de polymerase en chaine (PCR); 30% des échantillons contenaient des inhibiteurs de la PCR; un prélèvement sur 52 (1.9%) était positif pour la présence de M. leprae parmi les contacts des patients PB, et 13 sur 164 (7.9%) parmi les contacts de patients MB. Comme cette différence n'est pas significative et que certains cas de positivité ont été trouvés parmi les contacts de patients MB traites et guéris de leur infection, on en conclut que les infections observées ont été acquises à partir de la communauté.RESUMEN
Se estudiaron muestras de exudado nasal de 4 pacientes con lepra paucibacilar (PB), de 8 pacientes con lepra multibacilar y de sus contactos, para buscar la presencia de Mycobacterium leprae utilizando 2 ensayos de reacción en cadena de la polimerasa (PCR); 30% de las muestras contuvieron inhibidores de la PCR. Uno de 52 (1.9%) y 13 de 167 (7.9%) exudados resultaron positivos para M. leprae entre los contactos de los pacientes PB y MB, respectivamente. Puesto que esta diferencia no es significativa, además de que se encontraron algunos positivos entre los contactos de los pacientes MB tratados y curados de su infección, se concluye que las infecciones observadas se adquirieron de la comunidad.Although the route of infection by Mycobacterium leprae is unknown, infection through inhalation seems most probable in view of the large numbers of bacilli present in nasal secretions of lepromatous patients (1,3,7, 8).. In the absence of any possibility of the cultivation of M. leprae, studies on the presence of M. leprae in the nasal mucosa of contacts were based exclusively on Ziehl-Neelsen staining, with the inevitable shortcomings of lack of sensitivity and specificity.
The polymerase chain reaction (PCR) amplifying a fragment of DNA replaces to a great extent the cultivation of microorganisms. We undertook a study on the detection of a M. /eprac-specific DNA fragment in nose swabs from contacts of paucibacillary (PB) and multibacillary (MB) leprosy patients in Anjouan, Comores.
MATERIALS AND METHODS
Patients and contacts. Four PB and eight MB patients with clinically, bacteriologically, and histopathologically documented disease, as well as their household contacts (HHC), agreed to participate in the study. HHC were defined as persons sleeping during the night under the same roof. Patients and contacts were sampled during successive visits, during which the number of persons present was variable.
Nasal swabs. Nasal swabs were taken by introducing a cotton-tipped swab (VEL, Leuven, Belgium) 2-3 cm into each nostril successively, and rubbing gently on the lateral and median sides of each cavity. The swabs were kept in a cool place, sent to Antwerp by air, and stored at room temperature.
Specimen preparation. Sterile distilled water (0.5 ml) was introduced in each swab tube and vortexed with the swab in place for 1 min, then the swab was expressed against the tube wall and discarded. From each extraction fluid 15 µl was deposited on a multi-well slide, fixed, and stained by a cold Ziehl-Neelsen staining method (8). The 100 µg of extraction fluid was freeze-boiled five times (alternate passages through liquid nitrogen and boiling water bath) for 2 min and kept at - 20ºC until further processing. A negative control tube containing sterile distilled water was run in parallel with each sample.
The PCR technique I was performed as described previously (6): A final volume of 50 µl contained 10 µl of the sample; 10 pmoles of each primer (1); 200 µM each of dATP, dCTP, dGTP, dTTP; 1 unit of Taq polymerase (Cetus); 50 mM of KG; 10 mM of Tris-HG, pH 8.3; 2.5 mM MgCl2 and 100 µg of bovine serum albumin, and the internal control.
Amplification was done as follows: 94ºC, 2 min; 35 cycles of 55ºC, 2 min; 72ºC, 2 min and 94ºC, 1 min; followed by a final 72ºC, 10 min.
Amplicons. Amplicons were visualized by agarose gel electrophoresis and ethidium-bromide staining. Since a high number of nasal swab preparations contained inhibitors for the PCR as revealed by the absence of amplification of the internal control, all reactions were performed on the undiluted sample and a 1/10 dilution. If inhibitors were present in the 1/10 dilution, the sample was recorded as not interprétable.
The usual precautions to prevent contamination were taken, with the use of aerosolresistant tips (Biozym, Landgraaf, The Netherlands). Furthermore, each couple of reaction tubes was accompanied by and separated from the other samples by its negative distilled-water control. A series of nine nasal swabs taken from volunteers in Antwerp, Belgium, not involved in this work was also examined.
In order to confirm positive results, a PCR technique II was performed on all samples positive with the first technique using a second primer pair to amplify another region of the M. leprae genome. Primers complementary to a M. leprae-specific repetitive sequence were chosen for this purpose. However, the reaction conditions as described by Woods and Cole (9) were slightly modified, and the "hot start" technique was used to improve the yield and specificity of the reaction. PCR reactions were performed in a final volume of 50 µl containing 10 mM Tris HC1, pH 8.3; 50 mM KC1; 1 mM MgCK; 200 µM each of dATP, dGTP, dCTP; 400 µm dUTP; 10 pmol of each primer; 1 unit Taq polymerase and 5 µl sample. Instead of mineral oil, solid paraffin with a solidification point of 69ºC-73ºC (Merck) was used to overlay the reaction mix and a "hot start" realized as follows: after mixing all reaction components except Taq polymerase, about 50µl of melted paraffin was added to the aqueous layer and the test tube was heated for 10 min at 94ºC. After cooling to room temperature, 10 µl of a solution containing 0.1 unit Taq polymerase/µl in 10 mM Tris HC1, pH 8.3; 50 mM KC1 was added on top of the solidified paraffin.
After a first denaturation step at 94ºC for 3 min, 39 cycles of 1 min at 58ºC, 1 min at 72ºC and 1 min at 94ºC were performed with a final elongation time of 10 min.
The use of dUTP instead of dTTP allows reaction mixtures to be treated with uracilN-glycosylase (UNG; Perkin-Elmer) to exclude any possible contamination with the amplicon from previous reactions. To each tube 1 unit of UNG can be added before the addition of the paraffin, and the solutions incubated for 10 min at room temperature. The use of UNG fits nicely in the "hot start" procedure since the 10-min denaturation at 94ºC will cleave the dU-containing products and inactivate the enzyme simultaneously.
RESULTS
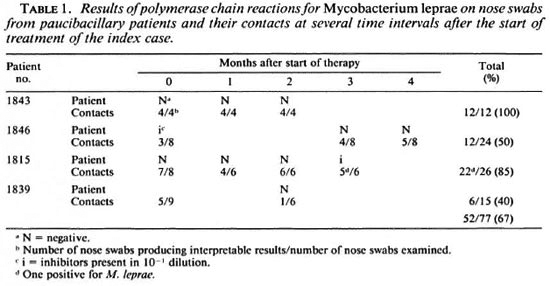
The nine nasal swabs from the Antwerp volunteers all produced negative results. PB patients and their contacts. Eleven nose swabs from 4 PB patients were examined (Table 1); 2 samples (patients 1846 and 1815) contained inhibitors at the 1/10 dilution, 9 samples were negative. Fifty-two of 77 (67%) nose swabs from contacts produced interpretable results. One sample (1.9%) was positive.
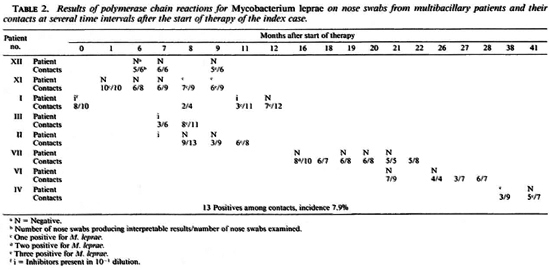
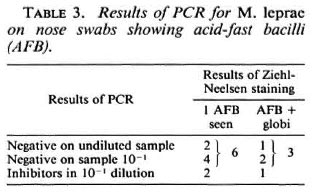
MB patients and their contacts. Twentythree nose swabs from eight MB patients were examined. Nineteen were interpretable, ofwhich three (from two patients, nos. XI and IV) were positive. One-hundred-sixty-four of the 236 (71%) nose swabs from contacts produced interpretable results; 13 (7.9%) were positive (Table 2). One contact person of patient VII produced a positive nose swab on two occasions, at 16 and 19 months after the start oftherapy ofthe index case. The sample taken at 18 months and those taken at 20, 21 and 22 months were negative. All other positives, when sampled before or after the positive result, were negative. However, eight positive nose swabs were among the last samplings done, and these persons were not examined at a later date. Table 3 shows a comparison between the PCR results for 12 cases where acid-fast bacilli (AFB) were seen in the corresponding smears. In 9 cases, 9 PCR results could be interpreted; 3 samples were PCR negative, 6 contained inhibitors when undiluted but were negative in the 10"1 dilution. In 6 cases only 1 AFB was seen; in 3 cases several AFB and globi were observed.
DISCUSSION
For more than a century the excretion of M. leprae through the nasal mucosa of MB patients has been documented, and it represents the most important portal ofexit for the organism. Whether M. leprae invades the human body after deposition of aerosolized organisms on the nasal mucosa is unknown. In the past, many attempts were made to detect M. leprae in Ziehl-Neelsenstained smears from nasal swabs, but this technique is always open to criticism, particularly because of its lack of specificity.
A species-specific PCR offers promise, in terms of both specificity and sensitivity.
We investigated the household contacts of a number of PB and MB leprosy patients. Sampling of the nasal mucosa through swabbing is not the optimal technique because it is impossible to standardize. However, under field conditions it is the only practical possibility; a nasal washing procedure as performed by Shepard (8) would be impossible.
The use of the previously described positive internal control in the PCR (6) has shown its importance since it revealed, in the present study, a high prevalence of inhibitors of the PCR. In 66 cases the inhibitory effect was absent in the 1/10 dilution, but in 30% ofthe samples it was still present in the 10-1 dilution, and these were scored as not interpretable. In future studies on nose swabs a nucleic-acid extraction method should be applied.
Only samples positive in the two different PCR techniques were taken into account. Nose swabs from two patients were positive; from one patient at months 8 and 9 after the start of treatment, and in a second patient 38 months after the start of therapy. These positives may represent residual M. leprae, present at the start of therapy and presumably killed by the therapy (the patients were treated with a combined drug treatment including rifampin and ofloxacin) or, particularly in the case positive at 38 months, new infections.
M. leprae was detected by PCR in 1.9% of 52 samples from contacts of PB patients and in 7.9% of 164 samples from contacts of MB patients. The difference is not significant (p = 0.20, two-tailed, Fisher's exact test). Among persons sampled repeatedly, only one (a contact of case VII) produced two positives, with an interval of 3 months, the sample taken between these two being negative. Three later samples taken from this person were also negative.
If the absence of a significant difference of positives between contacts of PB and MB patients can be confirmed, and on the basis of the present results, it seems that in a leprosy-endemic area most infections are community acquired. In Anjouan the case detection rate has remained stable at 0.38 per 1000 persons for the last 10 years (5).
Our results do not differ from those published by De Wit, et al. (4) who did not find a difference in the prevalence of M. leprae carriers in contacts and noncontacts of leprosy patients.
This investigation had to be stopped for reasons beyond our control. There is certainly a need to repeat and to extend it, also, in its technical aspect; for example, the need for alternative sample preparation in order to eliminate PCR inhibitors.
REFERENCES
1. Cox, R. A., KEMPSELL, K., FAIRCLOUGH, L. and COLSTON, M. J. The 16S ribosomal RNA of Mycobacterium leprae contains a unique sequence which can be used for identification by the polymerase chain reaction. J. Med. Microbiol. 35(1991)284-290.
2. BARTON, R. P. E. A clinical study of the nose in lepromatous leprosy. Lepr. Rev. 45(1974)135- 144.
3. D AVEY, T. F. and R EES, R. J. W. The nasal discharge in leprosy: clinical and bacteriological aspects. Lepr. Rev. 45(1974) 121-134.
4. DEWIT, M. Y. L., DOUGLAS, J. T., MCF ADDEN, J. and KLATSER, P. R. Polymerase chain reaction for detection of Mycobacterium leprae in nasal swab specimens. J. Clin. Microb. 31(1993)502-506.
5. PATTYN, S. R. and GRILLONE, S. Leprosy in the Comores 1981-88. Ann. Soc. Belg. Med. Trop. 71(1991)51-56.
6. PATTYN, S. R., URSI, D., IEVEN, M., RAES, V. and JAMET, P. Polymerase chain reaction amplifying DNA coding for species-specific rRNA of Mycobacterium leprae. Int. J. Lepr. 60(1992)234-243.
7. PEDLEY, J. C. The nasal mucous in leprosy. Lepr. Rev. 44(1973)33-35.
8. SHEPARD, C. C. Acid-fast bacilli in nasal excretions in leprosy and results of inoculation in mice. Am. J. Hyg. 71(1960)147-150.
9. WOODS, S. A. and COLE, S. A rapid method for the detection of potentially viable Mycobacterium leprae in human biopsies: a novel application of PCR. FEMS Microbiol. Lett. 65(1989)305-310.
1. M.D., Professor of Medical Microbiology, Institute for Tropical Medicine and University of Antwerp, Nationalestraat 155, B-2000 Antwerp, Belgium.
2. Dr.Sci.; University Hospital, Wilrijkstraat 10, B-2650 Edegem, Belgium.
3. Dr.Pharm.Sci., University Hospital, Wilrijkstraat 10, B-2650 Edegem, Belgium.
4.Dr.Anthr., Coordinator, Leprosy Control Anjouan, Mutsamudu, Anjouan, Federal Islamic Republic of the Comores.
5. Lab. Tech., Institute for Tropical Medicine, Antwerp, Belgium.
Received for publication on 4 January 1993.
Accepted for publication in revised form on 27 May 1993.