- Volume 58 , Number 1
- Page: 130–2
Further Evidence for the Exclusiveness of the Mycobacterium leprae-specific DNA Probe
To the Editor:
Sequence analysis of long reverse transcriptase generated stretches of the primary structure of 16S ribosomal ribonucleic acid (16S rRNA) from mycobacteria supported the phylogenetic position of Mycobacterium leprae within the subgroup of slow-growing pathogenic mycobacteria (7, 8). Based on sequence information, a 22-mer synthetic DNA oligonucleotide probe directed against a stretch of positions 206 to 227 (according to the IUB numbering system for Escherichia coli) was developed. The specificity of the DNA probe was tested in dot-blot hybridization using M. leprae, M. tuberculosis, M. avium, M. scrofulaceum, and M. phlei as reference organisms (10). Although the probe was exclusive for M. leprae, further experiments were needed to demonstrate the exclusive specificity of this probe for M. leprae by using a larger variety of Mycobacterium strains.
Armadillo-derived M. leprae were isolated from the liver and spleen tissue of experimentally infected nine-banded armadillos held in the Division of Laboratory Animal Science, Research Institute for Experimental Biology and Medicine, Borstel, Federal Republic of Germany. The animals were kept under conditions at which contamination with other mycobacteria was excluded (2). The identity and the purity of M. leprae were verified by negative growth on synthetic media (5), by positive DOPA oxidase reaction (6), and by decolorization with pyridine (4). Purification was performed as described earlier (1). The isolates were positive in an indirect immunofluorescence technique using M. leprae-specific monoclonal antibody against the phenolic glycolipid-I antigen of the organism (3). For the isolation of bulk RNA (7, 8), 39 strains of 36 species of Mycobacterium (The Table) were cultivated on Löwenstein-Jensen medium at an optimum growth temperature.
A 22-mer DNA oligonucleotide with the composition 5'ACTCCTGCACCGCAAAAAGCTT 3' (10) was 5'labeled with 32P γ-ATP and purified electrophoretically. Crude RNA (100 ng) from M. leprae and the other 39 mycobacterial strains were applied to Hybond N filters (Amersham) by a dot-blot apparatus (Schleicher & Schüll, Dassel) and fixed by UV 254 nm for 5 min. Prehybridization was in 6 x SSC and 4 x Denhardt's solution for 1 hr at 46ºC. Hybridization with the labeled probe (3 x 106 cpm, 30 ng) was performed in the same solution for 3 hr at 46ºC (20ºC below Tm of the probe). Subsequently, the filters were washed at 46ºC, 56ºC and 65ºC, each step for 15 min. Autoradiography was for 12 hr.
The Figure shows the results of dot-blot hybridization. At 46ºC (A in The Figure), all RNA dots gave a strong signal because under these washing conditions not only the homologous M. leprae rRNA but the heterologous rRNAs as well hybridized with the probe. To eliminate nonspecific binding the filters were washed at increased temperatures-at 56ºC and then at 65ºC, which corresponds to 1ºC below the melting point (Tm) of the homologous hybrid. Hybridization signals of the heterologous mycobacteria rRNA diminished due to the stringent conditions. Only a few strains still showed weak signals which are detectable as very light dots in B in The Figure. However, the homologous M. leprae rRNAprobe hybrid retains the distinct signal even at a temperature of 65ºC, which in its intensity is comparable to the signal obtained at optimal temperature of 46ºC.
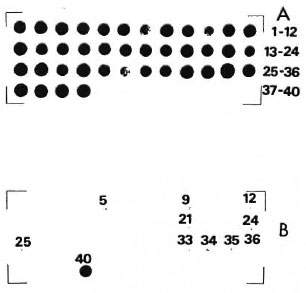
The Figure. Autoradiogram of a dot-blot hybridization between a M. leprae-specific oligonucleotide probe and bulk RNA from 40 strains of 37 Mycobacterium species. (Order of strains is as indicated in The Table.) A = nonstringent washing temperature (46ºC); B = stringent washing temperature (65ºC). Numbers (excepting No. 40, which is the homologous reaction) refer to those strains whose rRNA gave faint hybridization signals with the probe.
These results confirm the high specificity of the M. leprae probe. In addition to the information summarized in The Figure, we can also predict the failure of the probe to bind against the rRNA of M. triviale and M. fallax (Dorsch, Lévy-Frébault and Stackebrandt, unpublished). This is concluded from sufficiently large differences in the sequences of the target site of the two organisms.
In conclusion, the molecular-genetic technique facilitates identification of M. leprae using the species-specific probe as already discussed in a previous paper (10), when a very limited number of mycobac terial strains only was tested.
- Christian Pitulle, cand. Dipl. Biol.
Dagmar Witt, Dipl. Biol., cand. Ph.D.
- Erko Stackebrandt, Ph.D.
Professor
Institut für Allgemeine Mikrobiologie
Christian-Albrechts-Universität
Olshausenstrasse 40
2300 Kiel, Federal Republic of Germany
-Jindrich Kazda, Ph.D.
Priv. Doz.
Forschungsinstitut Borstel
Institut für Experimentalle Biologie und Medizin
2061 Borstel, Federal Republic of Germany
REFERENCES
1. DRAPER, P. Protocol 1/79: purification of M. leprae. In: Report of the Enlarged Steering Committee for Research on the Immunology of Leprosy (IMMLEP) Meeting of 7-8 February 1979. Geneva: World Health Organization, 1979, Annex 1, p. 4.
2. KAZDA, J. Nine-banded armadillos in captivity: prevention of losses due to parasitic diseases; some remarks on mycobacteria-free maintenance. Int. J. Lepr. 49(1981)345-346.
3. KOLK, A. H. J., Ho, M. R., KLATSER, P. R., EGGELTE, T. A., KUIJPER, S., DE JONGE, S. and VAN LEEUWEN, J. Production and characterization of monoclonal antibodies to Mycobacterium tuberculosis, M. bovis (BGG) and M. leprae. Clin. Exp. Immunol. 58(1984)511-521.
4. MCCORMICK, G. T. and SANCHEZ, R. M. Pyridine extractability of acid-fastness from M. leprae. Int. J. Lepr. 47(1979)495-499.
5. PORTAELS, S. F. and PATTYN, S. R. Isolation of fastidiously growing mycobacteria from armadillo livers infected with Mycobacterium leprae. Int. J. Lepr. 50(1982)370-374.
6. PRABHAKARAN, K. A rapid identification test for Mycobacterium leprae. Int. J. Lepr. 41(1973)121.
7. SMIDA, J. Reverse Transcriptase Sequenzierung von 16S rRNA: ein Beitrag zur Phylogenie der Ordnung Actinomycetales. Ph.D. thesis, Universit of Kiel, 1988.
8. SMIDA, J., STACKEBRANDT, E. and KAZDA, J. Molecular-genetic evidence for the relationship of Mycobacterium leprae to slow-growing pathogenic mycobacteria. Int. J. Lepr. 56(1988)449-454.
9. STACKEBRANDT, E. and CHARFREITAG, 0. Partial 16S rRNA primary structure of five Actinotiryces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J. Gen. Microbiol. (submitted).
10. STACKEBRANDT, E., SMIDA, J. and KAZDA, J. Theprimary structure of the 16S-rRNA of Mycobacterium leprae: its use in phylogeny and development of DNA probes. Acta Leprol. (Geneve) 7 Suppl. 1(1988) 222-225.
Reprint requests to Dr. Kazda.
We were deeply saddened to learn of the death of Dr. Chapman H. Binford at his home on the afternoon of 9 February 1990. Dr. Binford was an Honorary Vice President of the International Leprosy Association, and well known to generations of leprologists.
