- Volume 57 , Number 2
- Page: 465–71
Genetic susceptibility to leprosy on a Caribbean island: linkage analysis with five markers
ABSTRACT
Our recent segregation analysis, carried out on 27 large pedigrees f rom a Caribbean island (Desirade), has shown the presence of recessive major gene(s) controlling susceptibility to leprosy per se and nonlepromatous leprosy, respectively. Linkage analysis was performed between each of these two detected genes and each of five markers typed in the Desirade population: HLA, ABO, Rhesus, Gm and Km. No positive significant lod score was observed. However, for leprosy per se close linkage was excluded with Rhesus and Gm (and also with ABO and HLA, considering a lower value for the frequency of the gene controlling susceptibility to leprosy per se). The highest lod score, although not significant, was obtained between the gene for nonlepromatous leprosy and ABO. Our ovcrall rcsults, joined with previous studiesand experimental data, suggest that the gene controlling susceptibility to leprosy per se and that controlling susceptibility to nonlepromatous leprosy might be différent, acting at successive stages of the immunc response to infection with Mycobacterium leprae.RÉSUMÉ
Notre récente analyse de ségrégation menée sur 27 grandes généalogies d'une île antillaise (la Désirade) a mis en évidence l'existence de gènc(s) majeur(s) récessifs) contrôlant d'une part la susceptibilité à la lèpre per se et d'autre part la susceptibilité à la lèpre non lépromateuse. Une analyse de linkage a ici été réalisée, entre chacun de ces deux gènes détectés et chacun des 5 marqueurs qui ont été typés dans la population désiradienne: HLA, ABO, Rhésus, Gm et Km. Aucun linkage significatif n'a été observé. Cependant, pour la lèpre per se, un linkage étroit a été exclu avec Rhésus et Gm (et aussi avec ABO et HLA en considérant une fréquence plus basse pour le gène contrôlant la susceptibilité à la lèpre per se). Le lod score le plus élevé, bien que non significatif, a été obtenu entre le gène contrôlant la lèpre non lépromateuse et ABO. L'ensemble de nos résultats, combiné avec ceux des études précédentes et les données expérimentales, suggèrent que le gène contrôlant la susceptibilité à la lèpre per se et celui contrôlant la susceptibilité à la lèpre non lépromateuse pourraient être différents, intervenant à des étapes successives de la réponse immunitaire à l'infection par Mycobacterium leprae.RESUMEN
Un análisis de segregación reciente de 27 genealogías importantes de una isla del Caribe (Desirade) demonstró la presencia de gen(es) mayor(es) recesivo(s) que controlarían la susceptibilidad a la lepra per se, como así, a la lepra no lepromatosa. Sc efectuaron análisis de linkage entre cada uno de estos dos genes detectados y cada uno de los 5 marcadores determinados en la población de Desirade: HLA, ABO, Rhésus. Gm y Km. No se obtuvo linkage positivo. Sin embargo, se excluyo linkage próximo entre el gen de la lepra per se y Rhésus y Gm; el mismo resultado se obtuvo con H LA y ABO, considerando un valor menor de frequencia del gen que controlla la susceptibilidad a la lepra per se. El lod score mas alto, aunque no significativo, sc obtuvo entre el gen de la lepra no lepromatosa y ABO. Nuestros resultados, conjuntamente con aquellos de estudios previos y con los hallazgos experimentales, sugieren que el gen que controla la susceptibilidad a la lepra per se y aquel que controla la susceptibilidad a la lepra no lepromatosa podrían ser diferentes, interviniendo en etapas succesivas de la respuesta inmune a la infección por Mycobacterium leprae.Genetic susceptibility to leprosy is a widely studied field but, although there is now accumulating evidence for the important role of genetic factors in the host response to the etiologic agent, Mycobacterium leprae, the nature of this genetic component remains largely imprecise. One fact that emphasizes the genetic contribution to the expression of leprosy is the large individual variation in the response to M. leprae which is correlated with the cellular immune response of the patient (19). Whereas most infected individuals develop an effective immunity without disease, the others present a wide spectrum of clinical manifestations with, at one pole, tuberculoid patients displaying a positive, acquired cell-mediated immunity and, at the other pole, lepromatous patients showing an absence of specific T-cell response to M. leprae.
Animal models have allowed us to determine the genetic mechanisms of the host's ability to respond to mycobacterial infections. Experimental infections in mice have shown that the gene located on chromosome 1, which controls the natural resistance to different intracellular pathogens also regulates the distribution of the trait of resistance/susceptibility to M. lepraemurium among mouse strains (3,24). The acquired resistance, occurring at a later stage of the immune response and related to the specific cell-mediated response, seems to depend on more complex genetic mechanisms, among which the major histocompatibility complex (MHC) has been shown to play a role (5).
In humans, approaches to the genetic susceptibility to leprosy have progressed along two complementary lines-on one line are studies of familial aggregation of leprosy, including various investigations as twin studies (reviewed in 2,25) and, more recently, complex segregation analyses; on the other line are the relationships between leprosy and genetic markers, especially HLA, at either the population or the familial level. Segregation analysis is a statistical method used to determine from family data the mode of inheritance of a given trait, especially with the goal to elucidate single gene effects. Several segregation analyses already have been performed on leprosy but no definite conclusion could be reached. Whereas segregation of a recessive major gene for lepromatous (25) and nonlepromatous (9,11) leprosy respectively was proposed by some authors, it was not supported by others (21,22). Studies between leprosy and HLA have provided other sources of evidence for the role of genetic factors. At the population level, the most consistent results were found with the HLA class II antigens, principally HLA-DR2 (associated with tuberculoid leprosy and lepromatous leprosy) and HLADR3 which is associated with tuberculoid leprosy and decreased among lepromatous patients (reviewed in 17,26). Population studies with Gm immunoglobulin allotypes have recently shown a significant excess of some Gm haplotypes in lepromatous Vietnamese patients (6) and in nonlepromatous patients from Desirade, a Caribbean island (9).
Familial studies are of greater interest and allow one to demonstrate the genetic nature of the disease's relationship with the marker. Significant results have been obtained when studying the segregation of parental HLA haplotypes among affected siblings. A nonrandom segregation of parental HLA haplotypes among sets of tuberculoid leprosy children was observed in families from Surinam (7), India (10), and Venezuela (27). The same results were found among lepromatous leprosy siblings from Venezuela (27) and China (29). However, the observed random segregation of HLA haplotypes among all leprosy patients and among healthy siblings (26) in multicase leprosy families argued against the existence of HLA-linked factors playing a role in susceptibility to leprosy per se (17).
Another way to characterize a disease gene is to perform linkage analysis which allows one to test whether this susceptibility gene and a marker locus are located, more or less closely, on the same chromosome. Two previous linkage analyses limited to the study of the tuberculoid form of leprosy could not demonstrate a tight linkage between a susceptibility gene and the HLA system, and suggested a more complex genetic mechanism (11,20). Another linkage analysis, performed on 75 families from 11 different countries, showed the exclusion of close linkage between HLA and leprosy per se; from the same data, a tight linkage between HLA and in vivo response to lepromin was observed (8).
Our recent complex segregation analysis performed on 27 multigcncrational pedigrees from a Caribbean island (Desirade) has led to the detection of recessive major gene(s) controlling susceptibility to leprosy per se and nonlepromatous leprosy respectively (1). The aim of the present study is to perform linkage analysis between each of these two traits and any one of five markers typed in this population, HLA, ABO, Rhesus, Gm and Km, in an attempt to determine the location of these susceptibility genes and to assess whether or not they are different.
MATERIALS AND METHODS
Family data. The data and sampling procedure have been published elsewhere (1) and will be briefly summarized here. The pedigrees were ascertained through all leprosy patients living in Desirade, a small French West Indies island, in 1984. The accurate form of the disease was assigned to them according to present knowledge by using the available clinical, histological, bacteriological and immunological information. For the analysis, the patients were classified as either lepromatous-including the borderline lepromatous (BL) and the polar lepromatous (LL) forms of the Ridley and Jopling classification (18) -or nonlepromatous-including the polar tuberculoid (TT), the borderline tuberculoid (BT), and the borderline (BB) forms. The whole sample comprised 27 multigcncrational pedigrees with a total of 953 individuals including 82 leprosy patients (54 nonlepromatous and 28 lepromatous).
The pedigrees, including at least two affected subjects, informative for linkage, were selected from this sample. Table 1 presents the number of predigrees, informative matings, individuals and leprosy patients available for linkage analysis according to each marker.
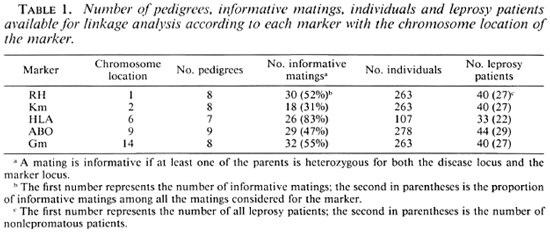
Marker determinations. Five markers were considered for the present linkage analysis: HLA, Gm and Km immunoglobulin allotypes, ABO and Rhesus (RH). Table 1 indicates the chromosomal assignment of each marker.
Typing of the HLA-A and HLA-B specificities was performed by means of the standard lymphocyte microlymphocytotoxicity technique with a set of 120 alio human anti-sera (14). HLA-DR typing was possible in only a few cases, but since the distance between the HLA-D and the HLA-B locus is less than 1% recombination, the recombination estimate between the D locus and the disease locus should be very similar to the one observed in this analysis.
Typing for Gm and Km immunoglobulin allotypes was carried out with the conventional agglutination inhibition technique (28). There are 10 Gm haplotypes (3 usually encountered in Europeans and 7 in Africans) which can account for the 37 observed phenotypes in Desiradians.
ABO and Rhesus determination was performed using classical techniques.
Genetic analyses. To evaluate the linkage relationship between two loci, the relative distance between two genes on the same chromosome is assumed to be related to the number of crossing-overs that occurs between them during mciosis. Because a crossing-over corresponds genetically to a recombination, an increased distance between two loci is expected to be associated with a higher recombination fraction, Θ. A Θ value of 0.5 corresponds to the independence between two loci (i.e., the two loci are on two different chromosomes or on the same chromosome but far enough apart so that numerous crossing-over events occur between them and their segregation is independent). The lod score method (17) is used to test linkage. It measures the odds of linkage for different recombination values (Θ = 0.0,..., 0.5) versus no linkage (Θ = 0.5). A lod score of 3.0 (1000 to 1 odds) is considered as significant evidence in favor of linkage and a score of -2.0 is taken as evidence against linkage at a given Θ value.
Linkage analysis assumes that the presence of the disease locus itself already has been suggested from other sources of evidence. Successive segregation analyses were previously performed (1), separately considering as affected a) all leprosy patients, b) nonlepromatous patients only, and c) lepromatous patients only. The results, summarized in Table 2, were consistent with the presence of a recessive major gene controlling susceptibility to leprosy per se and nonlepromatous leprosy, respectively. Two successive linkage analyses were then performed using the computer program LINK-AGE (13) between the five markers and a) the detected major gene controlling susceptibility to leprosy per se (LPS gene) and b) the detected major gene controlling susceptibility to nonlepromatous leprosy (NLL gene).
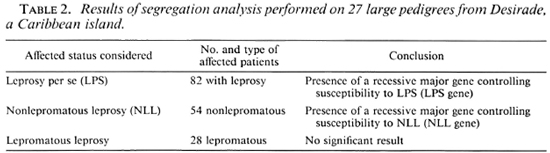
Linkage analysis needs to know some parameters at the disease locus: a) the frequency of the susceptibility allele, q, and b) the probability for an individual with a given genotype to express the disease phenotype (i.e., leprosy per se or nonlepromatous leprosy), or penetrance, which was considered to vary with age, categorized here into 10 classes. The maximum likelihood estimations of these parameters, taken from segregation analysis, were used for linkage analysis and are presented in Table 3. Since the method used in segregation analysis, breaking up the pedigrees into nuclear families, may have led to an overcstimation of q linkage analyses were repeated by dividing the q estimates by 10.
For ABO, Rhesus, Gm and Km, the allelic frequencies were estimated from the Desiradian population by maximum likelihood (results not shown). For HLA, if all the haplotypes are known, it is sufficient to take into account four "artificial" HLA alleles with equal frequencies in linkage analysis, as shown by Ott (16). In our sample, since HLA haplotypes are lacking for some individuals, it was necessary to consider five "artificial" alleles with equal frequencies.
RESULTS
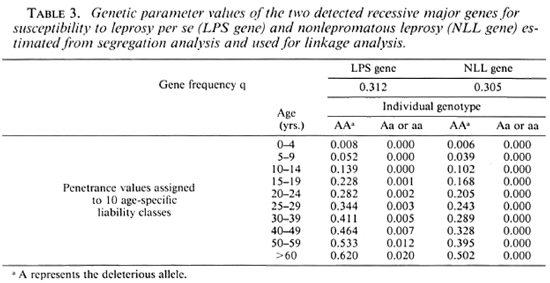
Results with leprosy per se. Table 4 presents the lod scores between the LPS gene and the five markers. No hint of linkage was discovered with these markers, the maximum lod scores all being close to 0. However, close linkage was excluded (z < - 2) for RH (at Θ < 0.01 or Θ < 0.10) and Gm (at Θ < 0.001 or Θ < 0.05) whether q or q/10 was considered. When q was divided by 10, tight linkage was also excluded for HLA and ABO.
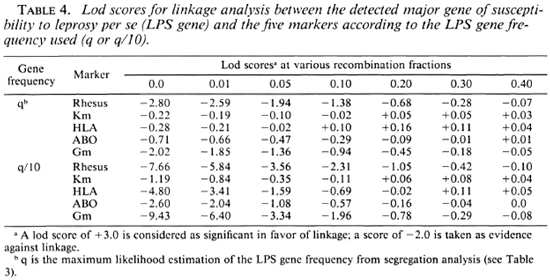
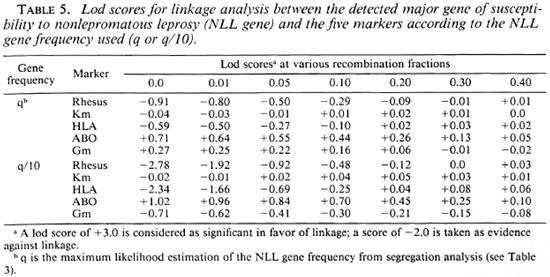
Results with nonlepromatous leprosy. Table 5 presents the lod scores between the NLL gene and the five markers. The highest lod score, although not significant, was observed for ABO with z = 0.71 (1.02 with q/10) at Θ = 0.0. When q was divided by 10, exclusion of tight linkage was obtained for RH (at Θ < 0.001) and HLA (at Θ < 0.001).
DISCUSSION
These linkage analyses do not show a significant linkage between either of the two susceptibility genes and the five markers typed in this Desirade population. For leprosy per se, close linkage was excluded with Rhesus and Gm. Results with ABO are of interest; whereas exclusion of tight linkage was observed for leprosy per se (when q was divided by 10), a positive maximum lod score of 0.71 (or 1.02 with q/10) was obtained at Θ = 0.0 for nonlepromatous leprosy. No previous linkage analysis has been performed with ABO and the results of population studies were not consistent, varying from the absence of association between ABO groups and leprosy to associations of either O, A or AB groups (2). Our finding needs further investigation to accumulate information and reach a significant conclusion. However, these opposite results with the same marker, as well as the previous observations in HLA family studies and experimental data, strengthen the hypothesis that the susceptibility genes controlling leprosy per se and nonlepromatous leprosy might be different. The observed exclusion of close linkage with HLA for leprosy per se (with q/10) is also in agreement with this last assumption and with the previous observations of a genetic factor controlling susceptibility to leprosy per se not linked to HLA. Although no positive and even negative lod scores were observed between HLA and nonlepromatous leprosy, these results are not in complete disagreement with the previous family studies demonstrating the role of HLA-linked factors in tuberculoid leprosy (7,10,27).
First, our familial data for HLA and non lepromatous leprosy might be not infor mative enough to detect a linkage between these two loci. Secondly, the methods used in these family studies, in contrast to linkage analysis, need no assumption on the underlying genetic model and our results show that a single HLA-linked gene cannot alone explain the familial distributions of nonlep romatous leprosy. Furthermore, if the sus ceptibility to a disease is controlled by two interacting genes (two-locus model), it has been shown that linkage analysis gives rise to erroneous and misleading results, showing frequent exclusion of tight linkage and a fairly large (15%-30%) estimation of the 0 value for the maximum lod scores (4,12). Besides, it could be seen that in the two previous linkage analyses limited to the tuberculoid form (11,20), the maximum lod scores were obtained for Θ values between 20% and 30%.
In conclusion, our whole genetic analyses in this Desirade sample, joined with the previous HLA family studies and experimental data, argue for a two-locus model of genetic susceptibility to leprosy. The detected major gene controlling susceptibility to leprosy per se might be an analog of the mouse natural-resistance gene acting at a first stage of the host response and is, in all probability, not linked to any of the five markers we have tested. The manifestation of leprosy forms then would be determined by at least another gene regulating later phases of the specific immune response. This genetic model might have important practical implications for prevention and treatment of leprosy. To confirm this hypothesis, we are planning segregation and linkage analyses using new models to take into account a two-locus mechanism, and a large survey in another population where epidemiological, biological and immunological data will be combined.
REFERENCES
1. ABEL, L. and DEMENAIS, F. Detection of major genes for susceptibility to leprosy and its subtypes in a Caribbean island: Desirade. Am. J. Hum. Genet. 42(1988)256-266.
2. BEIGUELMAN, B. An appraisal of genetic studies on leprosy. Acta Genet. Med. Gemellol. (Roma) 21(1972)21-52.
3. BROWN. I. N., GLYNN, A. A. and PLANT, J. Inbred mouse strain resistance to Mycobacterium lepraemurium follows the Ity/Lsh pattern. Immunology 47(1982)149-156.
4. CLERGET-DARPOUX, F. and B ONAITI-P ELLIE, C. Epistasis effect: an alternative to the hypothesis of linkage disequilibrium in HLA associated diseases. Ann. Hum. Genet. 44(1980)195-204.
5. CURTIS, J., ADU, H. O. and TURK, J. L. H2-linked genes which modify resistance of C57BL/10 mice to subcutaneous infection with Mycobacterium lepraemurium. Infect. Immun. 46(1984)635-638.
6. DE LANGE, G., WRIGHT P., VAN EEDE, P., VAN LEEUWEN, F., HOANG, T. L. and N GUYEN, T. D. Association between leprosy and immunoglobulin allotypes: Gm-A2m and Km frequencies in Vietnamese. J. Immunogenet. 11(1984)173-180.
7. DE VRIES, R. R. P., LAI A FAT, R. F. M., NIJENHUIS, L. E. and VAN ROOD. J. J. HLA-linked genetic control of host response to Mycobacterium leprae. Lancet 2(1976)1328-1330.
8. DE VRIES, R. R. P., S ERJEANTSON, S. W. and LAYRISSE. Z. Leprosy. In: Histocompatibility Testing 1984. Albert, E. D., Baur, M. P. and Mayr. W. R., eds. Berlin: Springer-Verlag. 1984, pp. 362-367.
9. DEMENAIS, F., GOULET, V., FEINGOLD, N., MILLAN, J., BLANC, M., RAFFOUX, C. and Bois, E. Genetic study of leprosy in a Caribbean island: Desirade. In: Genetic Control of Host Resistance to Infection and Malignancy. Skamenc, E., ed. New York: Alan R. Liss, 1985, pp. 319-324.
10. FINE, P. E. M., WOLF, E., PRITCHARD, J., WATSON, B., BRADLEY, D. J., FESTENSTEIN, H. and CHACKO, C. J. G. HLA-linked genes and leprosy: a family study in Karigiri, South India. J. Infect. Dis. 140(1979)152-161.
11. HAILE, R. W. C, ISELIUS, L., FINE, P. E. M. and MORTON, N. E. Segregation and linkage analysis of 72 leprosy pedigrees. Hum. Hered. 35(1985)43-52.
12. HODGE, S. and SPENCE, M. A. Some epistatic two-locus models of disease. II. The confounding of linkage and association. Am. J. Hum. Genet. 33(1981)396-406.
13. LATHROP, M. G., LALOUEL, J. M., JULIER, C. and OTT, J. Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am. J. Hum. Genet. 37(1985)482-498.
14. MITTAL, K. K. Standardization of the HLA typing method and reagents. Vox Sang. 34(1978)58-63.
15. MORTON, N. E. Sequential tests for the detection of linkage. Am. J. Hum. Genet. 7(1955)277-318.
16. OTT, J. A simple scheme for the analysis of HLA linkages in pedigrees. Ann. Hum. Genet. 42(1978)225-257.
17. OTTENHOFF, T. H. M. and DE VRIES, R. R. P. HLA class II immune response and suppression genes in leprosy. Int. J. Lepr. 55(1987)521-534.
18. RIDLEY, D. S. and JOPLING, W. H. Classification of leprosy according to immunity; a five-group system. Int. J. Lepr. 34(1966)255-273.
19. SANSONETTI, P. and LAGRANGE, P. H. The immunology of leprosy: speculations on the leprosy spectrum. Rev. Infect. Dis. 3(1981)422-469.
20. SERJEANTSON, S. W. HLA and susceptibility to leprosy. Immunol. Rev. 70(1983)89-112.
21. SERJEANTSON, S. W., WILSON, S. R. and KEATS, B. J. B. The genetics of leprosy. Ann. Hum. Biol. 6(1979)375-393.
22. SHIELDS, E. D., RUSSELL, D. A. and PERICAK-VANCE, M. A. Genetic epidemiology of the susceptibility to leprosy. J. Clin. Invest. 79(1987)1139-1143.
23. SKAMENE, E., GROS. P., FORGET, A., KONGSHAVN, P. A. L., ST. C HARLES, C. and TAYLOR, B. A. Genetic regulation of resistance to intracellular pathogens. Nature 297(1982)506-509.
24. SKAMENE, E., GROS. P., FORGET, A., PATEL, P. J. and NESBITT, M. N. Regulation of resistance to leprosy by chromosome 1 locus in the mouse. Immunogenetics 19(1984)117-124.
25. SMITH, D. G. The genetic hypothesis for susceptibility to lepromatous leprosy. Hum. Genet. 50(1979)163-177.
26. VAN E DEN, W. and DE VRIES. R. R. P. HLA and leprosy: a reevaluation. Lepr. Rev. 55(1984)89-104.
27. VAN EDEN. W., GONZALEZ. N. M., DE VRIES, R. R. P, CONVIT, J. and VAN ROOD, J. J. HLA-linked control of predisposition to lepromatous leprosy. J. Infect. Dis. 151(1985)9-14.
28. VYAS, G. N., FUDENBERG, H. H., PRETTY, H. M. and GOLD. E. R. A new rapid method for genetic typing of human immunoglobulins. J. Immunol. 100(1968)274-279.
29. Xu, K. Y., DE VRIES. R. R. P., FAI, H. M., VAN LEEUWEN, A., CHEN. R. B. and YE. G. Y. HLA-linked control of predisposition to lepromatous leprosy. Int. J. Lepr. 53(1985)56-63.
1. M.D.; Unite de Recherches de Génétique Epidemiologique, I.N.S.E.R.M., U. 155. Chateau de Longchamp, Bois de Boulogne 76016, France.
2. M.D.; Unite de Recherches de Génétique Epidemiologique, I.N.S.E.R.M., U. 155. Chateau de Longchamp, Bois de Boulogne 76016, France.
3. B.Sc.; Unite de Recherches de Génétique Epidemiologique, I.N.S.E.R.M., U. 155. Chateau de Longchamp, Bois de Boulogne 76016, France.
4. M.D.; Unite de Recherches de Génétique Epidemiologique, I.N.S.E.R.M., U. 155. Chateau de Longchamp, Bois de Boulogne 76016, France.
5. Ph.D., Unite de Recherches de Génétique Epidemiologique, I.N.S.E.R.M., U. 155. Chateau de Longchamp, Bois de Boulogne 76016, France.
6. Ph.D., Unite de Recherches de Génétique Epidemiologique, I.N.S.E.R.M., U. 155. Chateau de Longchamp, Bois de Boulogne 76016, France.
7. M.D., Centre d'Hemotypologie, C.N.R.S., Toulouse, France.
8. M.D., Centre National de Transfusion Sanguine. Paris. France.
9. M.D., Hôpital Saint-Louis, I.N.S.E.R.M., U. 93, Paris, France.
10. M.D., Institut Pasteur, Pointe a Pitre, Guadeloupe.
Reprint requests to Dr. Laurent Abel at his present address: Division of Biostatistics and Epidemiology, Howard University Cancer Center, 2041 Georgia Avenue NW. Washington, D.C. 20003. U.S.A.
Received for publication on 19 July 1988.
Accepted for publication in revised form on 8 December 1988.