- Volume 69 , Number 2
- Page: 99–103
No evidence of linkage between Mitsuda reaction and the NRAMP1 Locus
ABSTRACT
Thirty sib-pairs were ascertained through unrelated lepromatous probands. They consisted of 22 healthy individuals and 8 leprosy patients. The Mitsuda reactions of all sibs were evaluated both macroscopically and histologically, and high molecular weight genomic DNA was extracted f rom the white blood cells of all sib-pairs. Three DNA polymorphisms identified by polymerase chain reaction (274C/T, D543N, 1729 + 55del4) were used as chromosome markers at the NRAMP1 locus. Sib-pair comparisons did not disclose any sign of close linkage between the Mitsuda reaction and the genetic markers.RÉSUMÉ
Trente individus groupés en parents-descendants appariés ont été évalués à partir de cas-index (propositi) lépromateux sans liens de parenté. Ils consistaient en 22 individus cliniquement sains et 8 patients hanséniens. Les réactions de Mitsuda furent évaluées à la fois macroscopiquement et histolo- giquement, et de l'ADN génomique de haut poids moléculaire fut isolé des leucocytes de tous les individus groupés en parents-descendants appariés. Trois zones de polymorphisme de l'ADN identifiées par la réaction de polymerase en chaîne (274C/T, D543N, l729+55del4) furent utilisés comme marqueurs chromosomiques du locus nRAMPl. La comparaison des groupes de parents-descendants appariés n'a pas permis de révéler d'association étroite entre la réaction de Mitsuda et les marqueurs génétiques.RESUMEN
Se estudiaron Ia reacción de Mitsuda y los polimorfismos 274C/T, D543N y 1 729+55del4 en el locus RAMP1 en parejas de hermanos dentro de los cuales hubieron 22 indivíduos sanos y 8 pacientes con lepra. La reacción de Mitsuda se evaluó tanto macroscopicamente como histoiógicamente y los polimorfismos en cl DNA leucocitario, por la reacción en cadena de la DNA polimerasa. La comparación entre los hermanos no revelo ninguna asociación entre la reacción de Mitsuda y los marcadores genéticos estudiados.The intradermal injection of 0.1 lepromin may provoke a late response (Mitsuda reaction) which is a consequence of events that follow the phagocytosis of the lepromin's heat-killed Mycobacterium leprae by the skin macrophages (histiocytes). This late lepromin reaction may be clinically and histologically evaluated at 4 weeks. During the Sixth International Congress of Leprosy (31) it was recommended that live classes of the Mitsuda reaction should be clinically read as: absence of an observable or palpable element (negative reaction, -); a perceptible element smaller than 3 mm in diameter (doubtful reaction. ±); a conspicuous infiltrated element 3-5 mm in diameter (positive reaction, +); a conspicuous infiltrated element larger than 5 mm (positive reaction. ++); an ulcerated large nodule (positive reaction, +++).
When histologically evaluated, a positive Mitsuda reaction is defined by the presence of epithelioid cells, usually assuming a tuberculoid or tuberculoid-like structure, where acid-fast bacilli (AFB) are absent or scarcely found. This reaction indicates, therefore, that the macrophages are able to digest phagocytized heat-killed M. leprae, since this response is a consequence of the destruction of bacilli contained in lepromin by the macrophages which transform themselves into epithelioid elements. In the negative Mitsuda reaction the phagocytized AFB are not destroyed nor is a tendency to a tuberculoid structure seen (5, 6, 24). The association between the macrophages' ability to destroy the leprosy bacilli contained in lepromin microscopically observed and the clinical expression of this reaction is not complete. That is why in some cases histologic correspondence may not be found in either positive or negative Mitsuda reactions as read clinically (5, 6, 24).
Since the bacilli contained in lepromin are heat-killed, the Mitsuda reaction cannot be considered a replica of the leprosy infection. Nevertheless, this reaction has a high prognostic value, since Mitsuda-positive contacts of leprosy patients are free from the risk of manifesting lepromatous leprosy, which is completely associated with a negative Mitsuda reaction (15-25). Otherwise stated, a positive Mitsuda reaction indicates that the macrophages are able to destroy either dead or living leprosy bacilli. In contrast, leprosy contacts who persistently exhibit a negative Mitsuda reaction are considered to be at risk of contracting leprosy.
Early papers have shown that the macro- scopically evaluated Mitsuda reaction is a familial trait either among families free of leprosy (7, 9, 10) or among families of leprosy patients from Brazil (8) and from India (l9-28). Moreover, when the Mitsuda reaction was quantitatively analyzed, a significant parent-offspring correlation of the mean responses was observed in families free of leprosy as well as in families of leprosy patients (l2).
Since a higher proportion of Mitsuda- negative individuals is born to Mitsuda- negative parents, the opposite being observed in the offspring of Mitsuda-positive parents, this close parent-offspring association of the Mitsuda reaction favored the hypothesis that an autosomal gene pair could be responsible for this trait, the negative reaction being the recessive phenotype (7-9). This hypothesis was proven true recently when Feitosa, et al. (17) applied complex segregation analysis to family data of leprosy patients using the unified model of Lalouel, et al. (20). According to Feitosa, et al. (17). a Mendelian transmission for the Mitsuda reaction should be accepted beyond doubt, the positive phenotype being attributed to a partial dominant effect (degree of dominance estimated as 0.811).
These results stimulated the search for a locus responsible for the Mitsuda reaction. Taking into account that a) the Mitsuda- negative phenotype is associated with lep- romatous leprosy and b) the level of complexity of this reaction is obviously much lower than that required for the manifestation of this polar type of leprosy, the search for a locus responsible for the Mitsuda reaction seemed to be an easier way to find the locus responsible for susceptibility/resistance to lepromatous leprosy than the study of families of lepromatous patients (l6).
Thus, it was attractive to start linkage studies on the Mitsuda reaction with three polymorphic markers at the NRAMPl gene mapped in human chromosome 2q35 (21). This gene was proposed to be a major gene for susceptibility/resistance to mycobacterial infection since it is a homolog to the mouse Nrampl gene (natural resistance associated macrophage protein 1), formerly designed alternatively as Lsh, Ity or Bcg (11, 13, 32) in which a single amino acid change has been found to be associated with the inborn susceptibility to several intracellular parasites, such as M. lepraemurium, BCG, and species of Leishmania and Samonella (11).
SUBJECTS AND METHODS
Thirty sib-pairs were ascertained through unrelated lepromatous probands (index cases) living in Bauru in the state of Sao Paulo, Brazil, or in neighbor cities, and clinically examined by one of us (D.V.A.O.). The mean age of the probands was 44.5 ± 16.81 years, while the average age of their sibs was 42.6 ± 17.29 years. The macrophages' incapacity to destroy phagocytized M. leprae of the probands was demonstrated histopathologically and also by their clinically and microscopically examined Mitsuda-negative reactions. The probands' sibs consisted of 22 healthy individuals and 8 leprosy patients (5 lepromatous, 2 borderline bacilloscopically positive, 1 tuberculoid). The Mitsuda reactions of all these sibs were evaluated both macroscopically and histologically.
Since the late response to lepromin injection was evaluated both clinically and microscopically. the different degrees of posi- tivity of the Mitsuda reaction could be disdained, all individuals being able to be grouped for the present analysis in only two classes- as positive or negative Mitsuda reactors.
High molecular weight genomic DNA was extracted from the white blood cells of all of the sib-pairs according to standard protocols (29) and stored at 4°C in Tris- EDTA (10 mM Tris-HCl and 1 mM EDTA, pH 8.0). The three polymorphisms were identified by polymerase chain reaction (PCR) using primers described in Table 1 (21). The PCR amplification was carried out in 40 µl reaction volumes containing 50-100 ng genomic DNA, 10 mM Tris-HCL (pH 9.0), 50 mM KC1, 1.5 mM MgCl2, 0.2 mM of each dNTP, 20 pmol of each primer, and 1 unit of Taq polymerase (Pharmacia Biotech). After initial denaturation for 5 min at 94°C, amplification was accomplished using a protocol with 30 cycles including 1 min at 94°C, 1 min at 60°C and 1 min at 72°C, followed by a final extension of 10 min at 72°C in a MJ Research PT100 cycler. Negative controls were included in every run to test possible contamination.
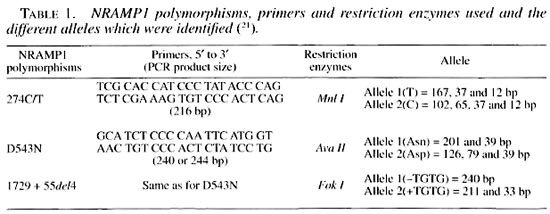
The genotyping of NRAMP1, 274 C/T polymorphisms using PCR produced a 216 bp fragment. Treated with Mnl I restriction enzyme at 37°C overnight, they defined the allele 1 (T) and allele 2 (C) of this polymorphism. Other PCRs were carried out for the determination of NRAMP1-D543N and 1729 + 55del4, using the same primers for both. The PCR products were either 240 or 244 bp fragments which, after being treated with Ava I or Fok I restriction enzymes, allowed the determination of the different alleles of the D543N and 1729 + 55del4 polymorphisms, respectively (Table 1). DNA fragments were resolved by electrophoresis in 12% acrylamide gels, ethidium bromide stained, and photographed under UV light.
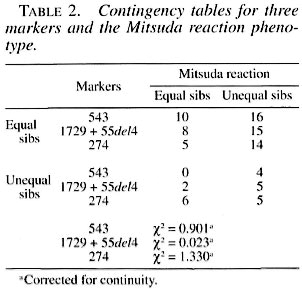
The unsophisticated formulation of Penrose (23) was applied to the data, and the tests of hypotheses were done by using chi- squared (χ2) in a two by two contingency table with one decree of freedom. The sib- pairs were classified as either equal or unequal for the marker, as for the Mitsuda phenotype.
RESULTS AND DISCUSSION
The results of these sib-pair comparisons are shown in Table 2, and they are clear in the sense that there is no evidence of linkage since the comparisons for these three markers were nonsignificant. The face value of the comparisons between the observed and expected values of the most informative marker, 274 C/T, which is a highly polymorphic locus, are in the opposite direction to those expected in the presence of linkage. In spite of dealing with a relatively small sample and using an unsophisticated statistical method, it should be kept in mind that when the same procedure was applied to this sample with regard to the markers used in this study, which are obviously known to be linked, 1 out of 3 tests gave the expected highly significant result (corrected yj = 21.970; 1 DF; p «0.001). These results agree with those of Shaw, et al. (30), Roger, et al. (26) and Roy, et al. (27), where no significant association could be found between the NRAMP1 locus and the different types of leprosy.
Some of the existing reported results require special discussion. Both in the Desi- rade Islands (14) and among the Vietnamese (') evidence of a major gene responsible for leprosy per se were observed, while this result could not be reproduced among the Chinese (-) or in a very large Brazilian sample (16). It is important to bring up this matter, in the present context, because these studies did not show any suggestion of the action of major genes in lepromatous leprosy, a disease which is highly correlated with the Mitsuda reaction.
Specifically, with respect to the genetics of the Mitsuda phenotype, there are also apparent discrepancies between the present Brazilian study and the results involving Chinese families reported by Abel, et al. (2) and the findings of Alca'is, et al. (4), pointing to a significant linkage between the Mitsuda phenotype and the NRAMP1 region.
Aside from the possibilities of statistical errors I and II on either type of studies, there is an agreement between the present findings and the very stimulating hypothesis of Roy, et al. (27) postulating the existence of at least two independent genetic mechanisms involved with this phenotype, lhe discrepancies being due to different combinations of frequencies of the loci in question. However, it should be borne in mind that there is an increasing amount of evidence linking the NRAMP1 locus to tuberculosis (8), also a macrophage-dependent infection. The seemingly agreement on the susceptibility to tuberculosis and NRAMP1 linkage on the one hand and the apparent heterogeneity of the data on leprosy on the other could be an indication of a greater complexity of the genetic mechanism involved in resistance/susceptibility to the latter.
Acknowledgment. The authors gratefully acknowledge the valuable support of FAPESP, CNPq and FAPERJ.
REFERENCES
1. Abel, L. and Demenais, F. Detection of major genes for susceptibility to leprosy and its subtypes in a Caribbesland: Desirade. Am. J. Hum. Genet. 42(1988)256-266.
2. Abel, L., Lap, N. D., Oberti. J., Thuc, N. V., Cua, V. V., Guilloid-Bataille, M., Schurr, E. and Lagrange, P. H. Complex segregation analysis of leprosy in Southern Vietnam. Genet. Epidemiol. 12(1995)63-82.
3. Abel. L., Sánchez. F. ()., Oberti, J., Thuc. N. V., Hoa, L. V., Lap, V. D., Skamene, E., Lagrange, P. H. and Schurr. E. Susceptibility to leprosy is linked to the human NRAMP1 gene. J. Infect. Dis. 177(1998)133-145.
4. Alcäis, A., Sánchez, F. O., Thuc, N. V., Lap. V. d., Oberti, J., Lagrange, P. H., Schurr. E. and Abel, L. Granulomatous reaction to intradermal injection of lepromin (Mitsuda reaction) is linked to the human NRAMPI gene in Vietnamese leprosy sibships. J. Infect. dts. 181(2000)302-308.
5. Azulay, R. D., Andrade, L. M. C, Silva. C., Rabello-Neto, A. V., Azulay, J. D., Garrido- Neves. R. and Miguez-Alonso, A. Comparison of the macroscopic readings and microscopic findings of the lepromin reaction. Int. J. Lepr. 28(1960)38-13.
6. Bechelli, L. M., Rath-De-Souza. P. and Quaglioto, R. Correlação entre os resultados da leitura clínica e do exame histopatológico da reação de Mitsuda. Rev. Bras. Lepr. 27(1959)172-182.
7. Beiguelman, B. Hereditariedade da reação de Mitsuda. Rev. Bras. Lepr. 30(1962)153-172.
8. Beiguelman. B. The genetics of resistance to leprosy. Int. J. Lepr. 33(Ï965)808-812.
9. Beiguelman. B. Lepromin reaction: genetic studies including twin pair analysis. Acta Leprol. 44(1971)5-65.
10. Beiguelman. B. and Quaglioto. R. Nature and familial character of the lepromin reactions. Int. J. Lepr. 33(1965)800-807.
11. Blackwell, J. The macrophage resistance gene Lsh/Ity/Bcg. Res. Immunol. 140(1989)767-828.
12. Botasso. Ò. A., Pou, H. O., Morini. J. C. and Rabasa, S. L. Estúdio genético de la resistência a la infección con el bacilo de Hansen en famílias con o sin lepra. Medicina (Buenos Aires) 44(1984)467-470.
13. Cellier, M. Govoni, G., Vidal, S., Kwan, T., Groulx. N., Liu. J., Sanchez, F., Skamene. E., Schurr, E. and Gros, P. Human natural resistance-associated macrophage protein: cDNA cloning, chromosomal mapping, genomic organization, and tissue specific expression. J. Exp. Med. 180(1994)1741-1752.
14. Demenais, E, Goulet, V., Feingold. N., Millan, J., Blanc, M., Raffoux, C. and Bois, E. Genetic study of leprosy in a Caribbean island: Desirade. In: Genetic Control of Host Resistance to Infection and Malignancy. Skamene, E., ed. New York: Alan R. Liss. 1985. pp. 319-324.
15. Dharmendra and Chatterjee, K. R. Prognostic value of the lepromin test in contacts of leprosy cases. Lepr. India 27(1955)149-152.
16. Feitosa, M. F., Borecki, I., Krieger, H., Beiguelman, B. and Rao, D. C. The genetic epidemiology of leprosy in a Brazilian population. Am. J. Hum. Genet. 56(1995)1179-1185.
17. Feitosa, M. F., Krieger, H., Borecki, I., Beiguelman, B. and Rao, D. C. Genetic epidemiology of the Mitsuda reaction in leprosy. Hum. Heredity 46(1996)32-35.
18. Greenwood, C. M. T., Fujiwara, T. M., Booth-royd, L. J., Miller, M. A., Frappier, D., Fanning. E. A., Schurr, E. and Morgan, K. Linkage of tuberculosis to chromosome 2q35 loci, including NRAMP1, in a large aboriginal Canadian family. Am. J. Hum. Genet. 67(2000)405-416.
19. Kundu, S. K., Ghosh, S., Hazra, S. K. and Chaudhury, S. Nature and familial character of lepromin sensitivity in 27 families and their siblings. Lepr. India 51(1979)465-474.
20. Lalouel. J. M., Rao, D. C., Morton, N. E. and Elston, R. C. A unified model for complex segregation analysis. Am. J. Hum. Genet. 35(1983):816-826.
21. Liu, J., Fujiwara, T. M., Buu, N. T., Sánchez, F. 0., Cellier, M., Paradis, A. J., Frappier, D., Skamene, E., Gros, P., Morgan, K. and Schurr, E. Identification of polymorphisms and sequence variants in the human homologue of the mouse natural resistance-associated macrophage protein gene. Am. J. Hum. Genet. 56(1995)845-853.
22. Michalany, N. S. and Michalany, J. Histopatologia da reação de Mitsuda em adultos sadios não comunicantes de hansenianos. Hansen. Int. 8(1983)105-123.
23. Penrose, L. S. The detection of autosomal linkage in data which consist of pairs of brothers and sisters of unspecified parentage. Ann. Eugen. 6(1935)133-138.
24. Petri, V., Mendes, E. V. and Beiguelman, B. Histology of the Mitsuda reaction of healthy adults with no known contacts with leprosy patients. Int. J. Lepr. 53(1985)540-545.
25. Quaglioto, R. Interpretação das reações limítrofes ou duvidosas do teste lepromínico. Boi. Serv. Nac. Lepra 21(1962)13-34.
26. Roger, M., Levee, G., Ciianteau, S., Gicquel, B. and Schurr, E. No evidence for linkage between leprosy susceptibility and the human natural resistance-associated macrophage protein 1 (NRAMP1) gene in French Polynesia. Int. J. Lepr. 65(1997)197-202.
27. Roy, S., Frodsham, A., Saha. B., Hazra, S. K., Mascie-Taylor, C. G. N. and Hill. A. V. S. Association of vitamin D receptor genotype with leprosy type. J. Infect. Dis. 179(1999)187-191.
28. Saha, K. and Agarwal. S. K. Immune deficit in patients with lepromatous leprosy: its nature and relation to genetic factors, spectrum, and duration of the illness. Int. J. Lepr. 47(1979)1-6.
29. Sambrook, J., Fritsch, F. and Maniatas, T. Molecular Cloning: a Laboratory Manual. 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory, 1989.
30. Shaw, M. A., Atkinson, S., Dockrell, H., Hussain, R., Lins-Lainson, Z., Shaw, J., Ramos, F., Silveira, F., Mehdi, S. Q., Kaukab, F., Khaliq, S., Chiang, T. and Blackwell, J. An RFLP map for 2q33-q37 from multicase mycobacterial and leishmanial disease families. No evidence for an Lsh/Ity/Bcg gene homologue influencing susceptibility to leprosy. Ann. Hum. Genet. 57(1993)251-271.
31. Sixth International Congress of Leprosy (Madrid). Immunology. The lepromin reaction. Int. J. Lepr. 21(1953)531-535.
32. Vidal, S. M., Malo. D., Vogan, K., Skamene, E. and Gros, P. Natural resistance in infection with intracellular parasites: isolation of a candidate for Bcg. Cell 73(1993)469-485.
1. Ph.D., Departmento de Genetica, Instituto Oswaldo Cruz. FIOCRUZ, Rio de Janeiro. Brazil.
2. M.D., Instituto Lauro de Sauza Lima, Bauru, SP, Brazil.
3. M.D., Instituto Lauro de Sauza Lima, Bauru, SP, Brazil.
4. Ph.D., Division of Biostatislics. Washington University School of Medicine, St. Louis. Missouri. U.S.A.
5. Ph.D., Departmento de Parasitologia, Instituto de Ciências Biomédicas, Universidade de Sao Paulo, Av. Lineu Prestes 1374, 05508-900 Sao Paulo, SP. Brazil.
6. Ph.D.. Departmento de Parasitologia, Instituto de Ciências Biomédicas, Universidade de Sao Paulo, Av. Lineu Prestes 1374, 05508-900 Sao Paulo, SP. Brazil.
Reprint requests to Dr. Beiguelman at the above address or FAX 56-11 -3818-7417; e-mail: bbeiguel@uol.com.br
Received for publication on 27 November 2000.
Accepted for publication on 5 April 2001.