- Volume 67 , Number 1
- Page: 52–9
Complete DNA sequence analysis for 16s Ribosomal RNA gene of the leproma-derived, cultivable and nerve-invading mycobacterium HI-75
ABSTRACT
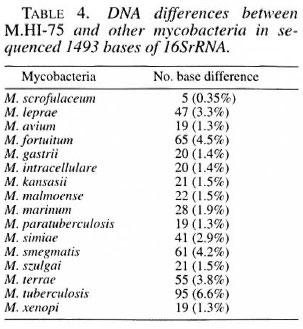
The complete 1493 nucleotide sequence of the 16SrRNA gene of the leproma-de- rived and cultivable mycobacterium HI-75 strain was analyzed to elucidate the taxonomic characteristics by direct sequencing of the polymerase chain reaction (PCR) products. The results revealed that the sequence of mycobacterium HI-75 was mostly similar to that of Mycobacterium scrofulaceum with 5 bases differences in the sequenced 1493 bases (0.35%) of the 16SrRNA gene. M. leprae differed f rom the strain with 47 bases (3.3%). Sasaki and Hamit reported the nerve-invasive activity of the inoculated mycobacterium HI-75 in nude mice or the l31 I-treated immunocompromised Swiss mice. The results indicate that mycobacterium HI-75 could be a mutant of M. scrofulaceum possessing the ability to invade the peripheral nerve in addition to developing leproma-like lesions.RÉSUMÉ
La séquence complète composée de 1493 nucléotides du gène du rARN (ARN ribosomal) de la sous unité 16 S provenant de la souche de mycobactérie HI-75 cultivable et dérivée d'un léprome, fut analysée pour déterminer les caractéristiques taxonomiques, par séquençage direct des produits de la réaction de polymérase en chaîne (PCR). Les résultats ont révélés que la séquence de la mycobactérie HI-75 était la plus proche de celle de Mycobacterium scrofulaceum . avec seulement 5 bases de différentes parmi les 1493 bases séquencées (0.35%) du gène 16SrRNA. Quarante sept bases (3.3%) séparaient M. leprae de la souche étudiée. Sasaki et Hamit ont rapporté la propriété neuro-invasive de la mycobactérie HI-75 chez les souris nues ou les souris Suisses immuno-déprimées traitées à l'iode 131. Les résultats indiquent que la mycobactérie HI-75 pourrait être un mutant de M. scrofulaceum possédant la propriété d'envahir les nerfs périphériques en plus de développer des lésions ressemblant aux lépromes.RESUMEN
Utilizando la técnica dc la reacción en cadena de la polimerasa (PCR) se analizó secuencia completa de los 1493 nucleótidos del gene 16SrRNA de la micobacte- ria HI-75, para dilucidar sus características taxonómicas. Esta bacteria no cultivable se aisló de un leproma de un paciente. Los resultados revelaron que la secuencia de la micobacteria HI-75 fue muy similar a la correspondiente a Mycobacterium scrofulaceum con solo 5 bases diferentes en la secuencia de 1493 bases (0.35%) del gene 16SrRNA. Por el contrario, la diferencia entre M. leprae y la micobacteria HI-75 fue de 47 bases (3.3%). Sasaki y Hamit reportaron que la bacteria HI-75 es capaz de invadir los nervios cuando se inocula en los ratones desnudos y en los ratones Suizos tratados con 131 I en inmunocomprometidos. Los resultados indican que la micobacteria HI-75 podria ser una mutante de M. scrofulaceum capaz de invadir los nervios periféricos y de inducir el desarrollo de lesiones lepromatosas.Although Hansen's disease is now under control in many countries, the causative organism Mycobacterium leprae still is regarded as a species of noncultivable bacilli. The aim of the present study is to ascertain the taxonomic characteristics of cultivable mycobacterium HI-75 (M.HI-75), which was isolated as Mycobacterium leprae by Skinsnes, et al. in 1975 from a lepromatous type of Hansen's disease patient (17 ). By the direct sequencing technique the 16S ribosomal RNA (16SrRNA) gene of the strain was analyzed in full length (1493 bases) and compared with those of other mycobacteria. The bacillus has been maintained in the laboratory of one of the authors since 1984 using Ogawa's medium enriched with glucuronic acid and N-acetyl- D-glucosamine. During that period of time, Sasaki and Hamit have observed both nerve invasion and the growth of inoculated M.HI-75 in either the nude mice or the 131 I-treated Swiss mice (9, 16).
MATERIALS AND METHODS
Bacterial strain. The leproma-derived and cultivable M.HI-75 has been maintained at 36.5°C utilizing either Ogawa's or Sauton's medium in the beginning and Ogawa's medium enriched with 1% each of glucuronic acid and N-acetyl-D-glucosamine which was recently used for this study.
Extraction of DNA. The M.HI-75 was suspended in 500 µl of TE buffer (10 mM Tris-HCl pH 7.6, 1 mM EDTA pH 8.0) to make a final suspension of 108 bacilli/ml and heated at 80°C for 20 min. Then, 50 µl of lysozyme (Wako Junyaku, Osaka, Japan) was added to the suspension at a rate of 10 mg/ml and the mixture was incubated at 37°C for 1 hr followed by mechanical disruption with glass beads for 1 min. The DNA was extracted using a Sepa-Gene kit (Sanko Junyaku, Tokyo, Japan) and finally precipitated with isopropanol. The precipitates were then washed with ethanol.
DNA amplification. A polymerase chain reaction (PCR) mixture containing 1 µl of 25 mM MgCl,, 10µl of 10x PCR buffer, 8µl of 2.5 mM each dNTPs, 60.5 µl of autoclaved ultra-pure water, 0.5 µl of Taq DNA polymerase (2.5 unit) (PCR kit; Takara, Ohtsu, Japan), 5 µl of each sense and antisense primers (20 pmole) (Table 1), and 5 µl of the DNA template mentioned above. The reaction was primed by a template melting step at 94°C for 5 min, followed by 35 cycles of the serial temperature changes consisting of 94°C for 30 sec, 55°C for 1 min and 72°C for 1 min using a Perkin-Elmer Model 480 DNA Thermal Cycler (Perkin-Elmer, Norwalk, Connecticut, U.S.A.).
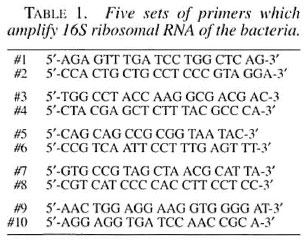
Direct sequencing technique. The PCR product thus obtained was directly sequenced by the use of an Auto Load Solid Phase Sequence kit (Amersham-Pharmacia Biotec, Uppsala, Sweden) including T7 DNA polymerase, biotinylated primers and Dye Amidite-667 conjugated primers (Amersham-Pharmacia Biotec, Tokyo, Japan) on an A.L.F.red sequencer (Amersham-Pharmacia Biotec, Uppsala, Sweden) at 1500V for 360 min on a 6% polyacrylamide gel with a 0.55 mm thickness. Both the sequences of sense and antisense strands were analyzed to confirm precision. The sequenced results thus obtained from M.HI-75 16SrRNA were compared with those of other mycobacteria using GenBank data or those by Edwards, et al . (6) on Rogall, et al . (14, 15).
RESULTS
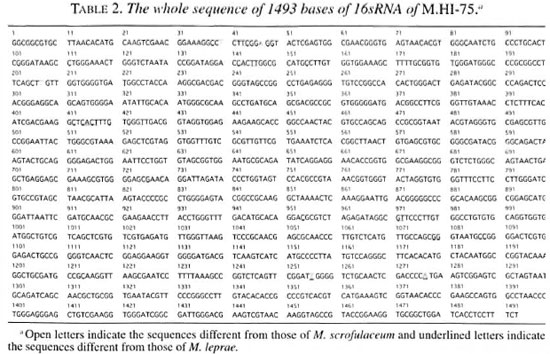
Table 2 shows all of the 1493 base sequences for the 16SrRNA gene obtained from the M.HI-75 strain. The sequences were compared with those of the other mycobacterial species shown in Table 3. Most of the sequences were stable in the 17 examined mycobacterial species. There were nine variable regions in the 16SrRNA gene of the bacteria. Several variable regions were also found between mycobacteria species, such as the region from nucleotide No38 to No50, from No 134 to No 178, and from No411 to No438. Three regions are shown in boxes (1, 2 and 3) in Table 3A. In the first region (No38-50), two substitute nucleotides were observed; however, these sequences are consistent with those of M. gastri , M. kansasii or M. simiae . But in the second and third regions, M.HI-75 shows no nucleotide difference from that of M. scrofulaceum . Other than these variable regions, M.HI- 75 has only three nucleotides that are different from that of M. scrofulaceum , but M. leprae shows 47 nucleotide differences from that of M.HI-75. All in all, the 16Sr- RNA sequence of M.HI-75 was most similar to that of M. scrofulaceum and differed from it in only 5 bases (0.35%) of the complete 1493 bases.

DISCUSSION
The applicability of 16SrRNA sequences for bacterial classification is well accepted (2, 6-8, 20) The 16SrRNA gene consists of genomically highly conserved primary sequences and has nine species-specific variable regions (2, 7, 8). The mycobacterial species can be distinguished and defined by the nucleotide structure of their 16SrRNA (1, 14, 15). PCR-mediated direct sequencing is a rapid and reliable method for obtaining the appropriate species-specific nucleotide sequence information. 16SrRNA sequencing allowed us to resolve the long-standing confusion with respect to the taxonomic differentiation within the M. avium-M. intracellulare complex. We examined the whole sequence of 16SrRNA of M.HI-75 and found that the sequences of 5 out of 1493 bases (0.35%) deviated from those of M. scrofulaceum and it was most similar to M. scrofulaceum . This result suggests that M.HI-75 is a variant of M. scrofulaceum . The postulated secondary structure of 16SrRNA is aligned to produce maximum base pairing between complementary segments (l2). The molecule appears to have four major domains of folding. This structure is analogous to the cloverleaf of visualization of a tRNA and contains many regions of self-complementarity, capable of forming double-helical segments. Even distantly related 16SrRNA sequences show that the potentially double-strand regions are highly conserved. In this study, bases No. 40 (T) and No. 48 (A), and bases No. 1256 (T) and No. 1277 (A) are compensatory mutations in a secondary structure of 16SrRNA so as to maintain base pairing. Base No. 207 (T), included in a small loop, is independent of base pairing. So nucleotidic changes found in M.HI-75 are thought to be two base pairs and one point mutations from M. scrofulaceum .
The mycobacterium examined in this study was originally isolated as M. leprae by Skinsnes, et al . in 1975 from a leproma of a lepromatous-type Hansen's disease patient and, therefore, named as M.HI-75 by them (17). Stanford, et al . (18) claimed this bacillus to be a variety of M. marianum (syn. scrofulaceum ) in 1977. Sasaki, et al . reported the nerve invasion and growth of the inoculated M.HI-75 into nude mice or the131 I-treated immunocompromised Swiss mice (16). This episode provoked a doubt that there should be a missing link of knowledge between leprosy and M. scrofulaceum since the nerve invasion of the bacilli had been regarded as a characteristic of M. leprae and not of M. scrofulaceum . In addition to that, the other genomic study on M.HI-75 performed by Williams, et al . (1994, personal communication) by restriction fragment length polymorphism of the PCR product suggested the resemblance of the bacilli as either M. smegmatis or M. fortuitum and not M. scrofulaceum . Hamit also reported the peripheral nerve invasion of M.HI-75 on inoculation into nude mice (9) besides forming lepromatous lesions with numerous macrophages which were loaded with the mycobacteria. Our present study suggested that M. scrofulaceum possibly possesses a pathogenetic activity in the peripheral nerve causing leprous-like lesions.
M. leprae has proved refractory to all attempts at in vitro culture. In many of the trials the claimed M. leprae have been denied to be so because of the possession of characteristics which the other cultivable bacilli have or the absence of those which are taken as characteristic for M. leprae . And among the isolated and denied candidates for M. leprae , M. scrofulaceum received the highest score in frequency (11). A number of laboratories have described procedures for the detection and identification of M. leprae by PCR (3-5,10, 13, l4). However, these have been applied to purified organisms, M. leprae in skin biopsy, or even tested on nerve biopsy to specific identification of M. leprae but no attempt has appeared in the literature to identify the whole pathogenetic organisms obtained from the site of the leprous neuropathy by the 16SrRNA sequence. Is there a possibility that M. scrofulaceum might cause nerve invasion and, therefore, nerve lesion? To perform a genomic study on the whole bacilli from infected lepromatous lesions of Hansen's disease patients using 16SrRNA might provide the manner with which to answer this question.
Acknowledgment. This work was presented at the 15th International Leprosy Congress, 7-12 September 1998, in Beijing, China. The authors are thankful to Ms. C. Yamamura (Protein & Nucleic Acid Analysis Unit, Joint Research Center, Kyorin University School of Medicine) for her skillful assistance in DNA sequencing.
REFERENCES
1. Boddinghaus, B., Rogall, T., Flohr, T., Blocker, H. and Bottger, E. C. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28(1990)1751-1759.
2. Brosius, J., Palmer, M. L., Kennedy, P. L. and Noller, H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli . Proc. Natl. Acad. Sei. U.S.A. 75( 1978)4801-4805.
3. Clark-Curtiss, J. E. and Docherty, M. A. A species-specific repetitive sequence in Mycobacterium leprae DNA. J. Infect. Dis. 159(1989)7-15.
4. Clark-Curtiss, J. E., Jacobs, W. R., Docherty, M. A., Ritchie, L. R. and Curtiss, R. III. Molecular analysis of DNA and construction of genomic libraries of Mycobacterium leprae . J. Bacteriol. 161(1985)1093-1102.
5. Clark-Curtiss, J. E. and Walsh, G. Conservation of genomic sequences among isolates of Mycobacterium leprae . J. Bacteriol. 171(1989)4844-4851.
6. Edwards, U., Rogall, T., Blokkr, H., Emde, M. and Bottger, E. C. Isolation and direct complete nucleotide determination of entire genes; characterization of a gene coding for 16S ribosomal RNA. Nucl. Acids Res. 17(1989)7843-7853.
7. Gray, M. W., Sankoff, D. and Cedargreen, R. J. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved core structure in small subunit ribosomal RNA. Nucl. Acids Res. 12(1984)5837-5852.
8. Greisen, K., Loeffelholz, M., Purohit, A. and Leong, D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32(1994)335-351.
9. Hamit, S. [An experimental nerve lesion simulating leprous neuropathy produced in the nude mice by the inoculation of a leproma-derived and cultivable Mycobacterium HI-75.] Jpn. J. Lepr. 65(1996)174-179.
10. Hartskeel, R. A., De Wit, M. Y. L. and Klatser, P. R. Polymerase chain reaction for the detection of Mycobacterium leprae . J. Gen. Microbiol. 135(1989)2357-2364.
11. Nakamura, M. [ Mycobacterium leprae and M. lepraemuriuin .] Tokyo: Tokai University Press, 1985.
12. Noller, H. F. tRNA-rRNA interactions and peptidyl transferase. FASEB J. 7(1993)87-89.
13. Pukaytis, B. B., Gelber, R. H. and Shinnick, T. M. Rapid and sensitive detection of Mycobacterium leprae using a nested-primer gene amplification assay. J. Clin. Microbiol. 28(1990)1913- 1917.
14. Rogall, T., Flohr, T. and Bottger, E. C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J. Gen. Microbiol. 136(1990)1915-1920.
15. Rogall, T., Wolters, J., Flohr, T. and Bottger, E. C. Towards a phylogeny and definition of species at the molecular level with the genus Mycobacterium . Int. J. Syst. Bacteriol. 40(1990)323-330.
16. Sasaki, K. [On the mice foot pads inoculation of a leproma-derived and cultivable mycobacterium.] Annual Report of National Tama Research Institute. 35th Issue. Tokyo: National Tama Research Institute, 1985.
17. Skinsnes, O. K., Matsuo, E., Chang, P. H. C. and Anderson, B. In vivo cultivation of leprosy bacilli on hyaluronic acid based medium. 1. Preliminary report. Int. J. Lepr. 43(1975)193-203.
18. Stanford, J. L., Bird, R. G., Carswell, J. W., Draper, P., Lowe, C., McDougall, A. C., Mclin- tyre, G., Patty. S. R. and Rees, R. J. W. A study of alleged leprosy bacilli strain HI-75. Int. J. Lepr. 45(1987)101-106.
19. Wichitwechkarn, J., Karnjan, S., Shuntawuttisettee, S., Sornprasit, C., Kampirapap, K. and Peerapakorn, S. Detection of Mycobacterium leprae infection by PCR. J. Clin. Microbiol. 33(1995)45-49.
20. Woese, C. R. Bacterial evolution. Microbiol. Rev. 51(1987)221-271.
1. M.D.. Assistant Professor; Chairman and Professor, Department of Biochemistry and Molecular Biology, Kyorin University School of Medicine, Shinkawa 6-20-2. Mitaka. Tokyo 181-8611, Japan.
2. M.D., Chairman and Professor, Department of Biochemistry and Molecular Biology, Kyorin University School of Medicine, Shinkawa 6-20-2. Mitaka. Tokyo 181-8611, Japan.
3. M.D., Director, Leprosy Research Center, National Institute of Infectious Diseases, Tokyo, Japan.
Reprint requests to Dr. Sakai at the above address or FAX 81-422-407281.
Received for publication on 7 October 1998. Accepted for publication in revised form on 20 November 1998.