- Volume 68 , Number 2
- Page: 121–8
Mycobacterium leprae typing by genomic diversity and global distribution of genotypes
ABSTRACT
The genetic diversity and related global distribution of 51 Mycobacterium leprae isolates were studied. Isolates were obtained f rom leprosy patients f rom 12 geographically distinct regions of the world and two were obtained f rom nonhuman sources. Polymerase chain reaction (PCR) followed by DNA sequencing was performed targeting the rpoT gene of M. leprae. Isolates were classified into two groups based on the number of tandem repeats composed of 6 base pairs in the rpoT gene. Isolates f rom Japan (except Okinawa) and Korea belonged to one group, while those f rom Southeast Asian countries, Brazil, Haiti and Okinawa in Japan belonged to a second genotype. M. leprae obtained f rom two nonhuman sources (an armadillo and a mangabey monkey) revealed the latter genotype. These results demonstrate the genetic diversity of M. leprae and the related genotype-specific distribution in the world.RÉSUMÉ
La diversité génétique et la distribution géographique globale correspondante de 51 isolais de Mycobacterium leprae furent étudiées. Les isolats furent obtenus à partir de patients lépreux provenant de 12 régions géographiquement distinctes et deux isolats furent obtenus de source non-humaine. La réaction de polymerase en chain (PCR) suivie par le séquençage des produits obtenus fut réalisée sur le gène rpoT de M. leprae. Les isolats furent classés en deux groupes basés sur le nombre de motifs répétés 2 par 2 (tandem repeats) composés de 6 paires de bases au sein du gène rpoT. Les isolats du Japon (excepté Okinawa) et Corée appartenaient à un groupe, alors que ceux provenant de l'Asie du Sud-Est, du Brézil, d'Haïti et d'Okinawa au Japon appartenaient au second genotype. M. leprae provenant des deux sources non humaines (un tatou à neuf bandes et un signe mangabey) révélèrent ce dernier génotype. Ces résultats démontrent la diversité génétique de M. leprae et la distribution mondiale reliée au génotypes spécifiquement étudiés ici.RESUMEN
Se estúdio la diversidad genctica y la distribution global de 51 aislados de Mycobacterium leprae. Los aislados se obtuvieron de pacientes con lepra de 12 regiones geográficas distintas del mundo y de dos fuentes no humanas. El análisis se hizo por la reacción en cadena de la polimerasa (PCR) y por secuenciación del DNA de la region correspondiente al gene rpo T de M. leprae. Los aislados se clasificaron en dos grupos con base en el número de "secuencias tandem" compuestas por 6 pares de bases en el gene rpo T. Los aislados de Japón (excepto Okinawa) y Korea correspondieron a un genotipo, mientras que los aislados de los países del sudeste asiático, Brazil, Haiti, y Okinawa en Japon, correspondieron a un segundo genotipo. Los aislados de M. leprae obtenidos de dos fuentes no humanas (un armadillo y un mono mangabey) mostraron el sugundo genotipo. Estos resultados muestran la diversidad genética de M. leprae y la distribution de los genotips de M. leprae en el mundo.Leprosy is a chronic infectious disease caused by Mycobacterium leprae and is still a major health problem in the developing countries of Asia, Latin America and Africa (28). Global efforts to control leprosy by intensive chemotherapy have led to a significant decrease in the number of registered patients; however, new cases reported annually remain more than 0.5 million. The successful elimination of leprosy will likely depend upon a multifactorial approach, including appropriate chemotherapy, immunoprophylaxis and chemoprophylaxis. Critical to the success of any elimination strategy will be the identification and removal of the natural source of transmission and infection.
It has been difficult to identify sources of infection because of the protracted incubation period preceding clinical disease and because M. leprae remains uncultivable on artificial media. These two factors have slowed our understanding of the route of transmission of M. leprae and have continued to confound efforts to design strategies effective in eradicating the natural reservoir of M. leprae which could lead to the global elimination of this disease. Strain-specific markers could provide the necessary tools for understanding this aspect of the epidemiology of leprosy.
M. leprae appears to be minimally related to other mycobacteria based on the content of guanine plus cytosine, genome size (3- 16), unique biochemical features, such as phenolic glycolipid-I (15) and the presence of glycine in place of L-alanine in the peptide portion of peptidoglycan (l2). Comparative studies of M. leprae isolates by total genomic hybridization have shown significant homology, suggesting genetic similarity among them (2). Related studies of the M. leprae genome using restrietion fragment-length polymorphism (RFLP) have demonstrated that less than 0.3% of the nucleotides differed among the genomes in isolates of M. leprae from diverse origins, including leprosy patients, armadillos and mangabey monkeys (4). Williams, et al. reported that M. leprae isolates obtained from geographically distinct areas did not exhibit genotypic diversity by RFLP (27). de Wit and Klatser reported that isolates of M. leprae from different sources had identical 16S-23S rDNA intergenic spacer regions (9), a documented source of species and strain variability in some bacteria (14, 18, 19). In contrast, Fsihi and Cole demonstrated variability associated with the polA locus of M. leprae among eight chromosomal DNA samples originating from India, Africa and France (13). However, it was not clear whether diversity in the polA locus was applicable for typing of M. leprae.
Shepard and McRae have observed that the growth rate of M. leprae in mouse foot pads varies among isolates and that the differences are stable (24). Other studies have supported these results although the basis for phenotypes from the aspect of growth rate has not been elucidated. In an attempt to link growth rate with genotypic variation, we compared DNA sequences of various genes including rpoT, a homolog of the mycobacterial principal sigma factor (5, 11, 23). We found that a short repetitive region of rpoT exhibited a sequence polymorphism among isolates.
This report describes genetic diversity of the rpoT gene of M. leprae among 51 isolates obtained from 12 geographically distinct areas of the world and the distribution of genotypes.
MATERIALS AND METHODS
Source of M. leprae strains and preparation of genomic DNA. A total of 51 M. leprae isolates was used in this experiment. They were obtained from different areas of the world, and included clinical isolates from a biopsy specimen (BS) and those established by passages in nude mice (NM) or armadillos (AL). The countries from which M. leprae isolates were obtained and the number of isolates tested are as follows: Bangladesh (3 BS), Brazil (3 BS), Haiti (1 BS), India (1 AL). Indonesia (4 BS + 1 NM), Japanese Mainland (X BS + 4 NM), Japan Okinawa (2 BS + 3 NM), Korea (11 BS), Nepal (1 BS), Pakistan (1 BS), The Philippines (1 BS + 1 AL) and Thailand (3 NM + 1 AL). Two of the M. leprae isolates were from the nonhuman origins of a naturally infected armadillo (Louisiana, U.S.A; 1 AL) and a mangabey monkey (Nigeria; 1 AL).
M. leprae was purified from tissues as described (10). Extraction of chromosomal DNA was carried out as previously described (8). Biopsy samples were homogenized, and partially purified bacilli were disrupted with freezing and thawing followed by phenol extraction and ethanol precipitation. For the preparation of bacterial DNA from biopsy specimens embedded in paraffin, three to live sections (5 µm thick) were treated by the method of de Wit, et al. (8). The skin-scraped materials were suspended in phosphate-buffered saline (PBS), purified partially by centrifugation, and DNA was extracted after freezing and thawing.
PCR and sequencing. PCR was carried out using Ampli Taq DNA polymerase (Perkin-Elmer Applied Biosystems, Nor- walk, Connecticut, U.S.A.) in a 50 µl volume containing 150 pg of genomic DNA or 5 µl of template DNA solution and 1 µM of primers with reagents and protocols supplied by the manufacturer. Primers A (5'- AAGCGTCGATACAAAGGCACCGT-3') and B (5'-AGTAGCTTCGCCATCCTCGGTTT-3') were used for amplification to span the 300 bp fragment of the target region of the rpoT gene. The amplification was carried out in a thermal cycler (Astech PC800; Astech Co., Fukuoka, Japan) under the conditions of 30 sec at 95°C, 2 min at 60°C, and 4 min at 72°C for 30 cycles. The full length of the rpoT gene with up- and downstream regions was amplified with primers C (5'-GCTGTCGGTCACGGCTAT-3') and D (5'-GAAAACCGCACCCCGATGGT-3') under the conditions of 30 sec at 95°C, 2 min at 48°C, and 4 min at 72°C for 35 cycles. The short fragments, 91 bp or 97 bp, containing the target region of rpoT were obtained using primers E (5'-ATGCC- GAACCGGACCTCGACGTTGA-3') and F (5'-TCGTCTTCGAGGTCGTCGAGA- 3') under the same conditions as for the 300 bp fragment of the rpoT gene, except for 45 cycles of the thermal reaction. The same sequence information was obtained when EX Taq DNA polymerase (Takara Shuzo, Shiga, Japan), as 3'-5' exonuclease activity, was used for amplification.
DNA samples for sequencing were recovered from agarose gel after electrophoresis using an Easy Trap DNA purification kit (Takara Shuzo). Determination of sequences was performed in both directions using a Thermo Sequenase kit (Amersham Life Science, Cleveland, Ohio, U.S.A.) with 35S-dCTP or a BigDye Terminator Cycle Sequencing FS Ready Reaction kit (Perkin-Elmer) in an ABI Prism. 310 Genetic Analyzer (Perkin-Elmer). The nucleotide sequences obtained were analyzed using the DNASIS computer program (Hitachi Software Engineering, Yokohama, Japan).
Agarose gel electrophoresis. For comparing the differences in the repetitive region of the rpoT gene, the short fragments of the target region were amplified, and reaction products, 97 and 91 base pairs, were electrophoresed in a 4% Meta PhorTM agarose gel (FMC Bioproducts, Rockland, Maine, U.S.A.) using TBE buffer at 75 volts.
RESULTS
Sequence variation in the rpoT gene of M. leprae. DNA sequences of various genes involved in replication, transcription and translation, such as the rpoT, the rpoA, the tuf, the polA and the 16S rRNA genes, were studied to determine whether DNA polymorphism exists in these genes that might explain growth rate differences in the mouse foot pad. A single sequence variation in the rpoT gene was observed among isolates but it did not appear to differentiate fast- and slow-growing phenotypes. Alignment of the 300 bp target region of the rpoT gene showed that the number of tandem repeats, composed of 6 bp repeats (GACATC), was different among the isolates. One group of isolates contained three tandem repeats; whereas a second group contained four repeats in the target region of rpoT (Fig. 1). Origins of M. leprae isolates from the latter group were limited to Japan (except Okinawa) and Korea, while isolates with three tandem repeats were found in the Okinawa Islands, southern Asian countries such as Bangladesh, India, Indonesia, Nepal, Pakistan, The Philippines and Thailand, and the Americas (Brazil and Haiti). In addition, two nonhuman isolates of M. leprae (armadillo and mangabey monkey) showed the three tandem repeat genotype.
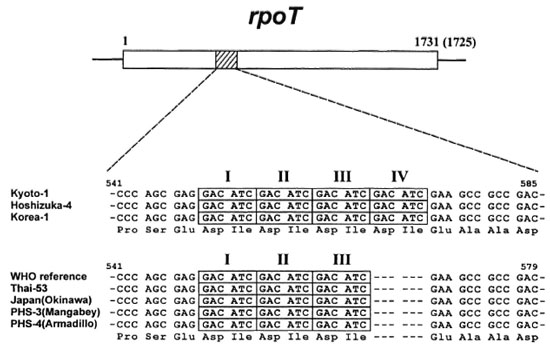
Fig. 1. Sequence alignment of DNA fragment corresponding from 541 to 585 (for 4 tandem repeats) or 541 to 579 (for 3 tandem repeats) of the rpoT gene from M. leprae isolates. Sequences from seven isolates and the reference (from Database, accession no. U15181) are shown. The nucleotide sequence data will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession numbers AB019193 and AB019194.
Comparative analysis of the complete rpoT DNA sequence (1725 bp coding for 575 amino acids) among isolates of both genotypes showed that only the 6 bp tandem repeats were different. This analysis also included up- and downstream regions for a total of 1900 bp. To confirm that the strains used in this study were M. leprae, we identified the M. leprae-specific fragment of the groEL gene by the method of Plikaytis, et al. (22).
Genotype detection by electrophoresis in agarose gel. To simplify the detection of the two groups, we developed an assay capable of classifying the two genotypes. The assay was based on amplifying an approximately 90 bp region containing the rpoT target region and analyzing the mobility of resultant amplicons on 4% Meta PhorTM agarose. The DNA fragments obtained by PCR from two genotypes were clearly separated as shown in Figure 2.
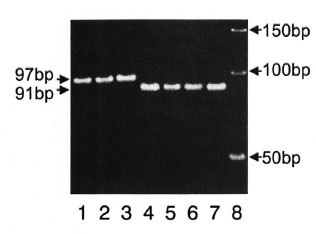
Fig. 2. Genotype detection by electrophoresis in agarose. Five (µl of PCR products were electro- phoresed in 4Vr Meta PhorTM agarose and then stained with ethidium bromide. Samples were: lane I, Kago- shima (Japan); lane 2, Kanazawa (Japan); lane 3, Ko- rea-1; lane 4. Thai-53; lane 5, Okinawa-1 (Japan); lane 6, PHS-3 (mangabey monkey); lane 7. PHS-4 (armadillo); and lane 8, the DNA size marker of 50 bp ladder.
DISCUSSION
The most striking finding was the apparent linkage of the four tandem repeat genotype between M. leprae strains isolated in Japan, excluding Okinawa, and those strains from Korea (Fig. 3A). The reason for this bias in distribution of the genotype with the four tandem repeat is not clear at present. A similar bias in genotype-specific distribution has been reported in Helicobacter pylori within Korean and Japanese populations. Epidemiological studies on H. pylori have indicated that almost all isolates from Korea and Japan are associated with the cag+ genotype; whereas in other parts of the world 30% to 40% of H. pylori strains are cag- (6- l7-21). In addition, H. pylori strains isolated from Asia could be separated from those of Caucasian origins on the basis of the nucleotide sequence (1, 25). It has been postulated that an H. pylori genotype-specific distribution is associated with human migration (7). Accordingly, it is reasonable to assume that the M. leprae strain- specific distribution that we observed may be related to patterns of human migration. Similarly, predominance of a single genotype was reported in M. tuberculosis in East Asian countries (26).
The M. leprae strains found in Okinawa were unrelated to the strains in the rest of the Japanese islands (Fig. 3A). There is a long history of concentrated interchanges between Korea and the main part of Japan, since the earlier migration of Mongolian people from Korea to Japan in the Yayoi and Kofun eras (300 BC to 600 AD). Several genetic markers have shown the close relationship between populations in Korea and most of Japan (20). In contrast, Okinawa was an independent country until the 19th century and had closer ties with southern Asian countries than with Japan.
The ubiquitous distribution of the three tandem repeat (Fig. 3B) suggests that further classification of this genotype will be necessary to distinguish strains of M. leprae for epidemiological studies. Fsihi and Cole reported genomic variability in a polA locus among five clinical isolates and three armadillo-grown strains of M. leprae (13). We examined this marker using eight strains of M. leprae (four strains with each rpoT type) according to their method but detected only one type of polA.
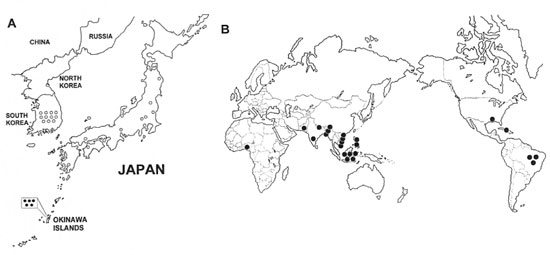
Fig. 3. Distribution of two genotypes of M. leprae isolates in Japan and Korea (A) and in the world (B).  = M. leprae with three repeats in the rpoT gene;
= M. leprae with three repeats in the rpoT gene;  = M. leprae with four repeats in the rpoT gene.
= M. leprae with four repeats in the rpoT gene.
Despite efforts to detect genomic diversity among M. leprae isolates, clearly defined genetic polymorphism useful for epidemiological purposes has not been reported. Our results demonstrate the genetic diversity of M. leprae based upon differences in the rpoT gene of 51 isolates. Strains of M. leprae isolated were divided into two genotypes and their distribution in the world showed a striking bias, suggesting a relationship to the movement of the Korean and Japanese people. Further analysis of M. leprae strains is needed to explain the bias seen in the global distribution as well as to appreciate the utility associated with rpoT typing M. leprae for epidemiological survey.
Acknowledgment. We thank Drs. Eiji Nagao (National Sanatorium Oshima Seishoen, Kagawa, Japan), Kunihiro Kinjoh (National Sanatorium Okinawa Airakuen, Okinawa. Japan), Masako Namisato (National Sanatorium Tama Zenshoen, Tokyo. Japan), Masamichi Goto (National Sanatorium Hoshizuka Keiaien, Kagoshima, Japan), Atsushi Hosokawa (School of Medicine, University of the Ryukyus, Okinawa, Japan), Akiko Obara (Medical College, Kyoto University. Kyoto. Japan), M. M. Bari (Leprosy Control Institute and Hospital, Dhaka, Bangladesh), L. E. Takaoka (Sociedate Filantrópica Humanitas, Brazil) for supplying clinical samples and Dr. M. J. Colston for supplying armadillo-grown M. leprae of an Indian isolate. We also express thanks to Mr. Kunio Kawatsu for preparing sections. This work was supported by a Health Science Research Grant of Research on Emerging and Re-emerging Infectious Diseases. Ohyama Health Foundation, Sasakawa Memorial Health Foundation. and the U.S.-Japan Cooperative Medical Science Program.
REFERENCES
1. Achtman, M., Azuma, T., Berg, D. E., Ito, Y., Morelli, G.. Pan, Z. J., Suerhaum. S., Thompson, S. A., van der Ende, A. and van Doorn. L.-J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32 (1999) 459-170.
2. Athwal, R. S., Deo, S. S. and Imaeda, T. Deoxyribonucleic acid relatedness among Mycobacteritim leprae. Mycobacterium lepraemurium and selected bacteria by dot blot and spectrophotometric deoxyribonucleic acid hybridization assays. Int. J. Sysl. Bacteriol. 34 (1984) 371-375.
3. Clark-Curtiss J. E.. Jacobs W. R., Docherty M. A.. Ritchie I.. R. and Curtiss. R., III. Molecular analysis of DNA and construction of genomic libraries of Mycobacterium leprae. J. Bacteriol. 161 (1985)1093-1102.
4. Clark-Curtiss. J. E. and Walsh, G. P. Conservation of genomic sequences among isolates of Mycobacterium leprae. J. Bacteriol. 171 (1989) 4844-4851.
5. Collins, D. M.. Kawakami, R. P., de Lisle, G. W.. Pascopella, L., Bloom, B. R. and Jacobs, W. R.. Jr. Mutation of the principal σ factor causes loss of virulence in a strain of the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U.S.A. 92 (1995) 8036-8040.
6. Covacci, A.. Censini, S., Bugnoli, M., Petracca, R., Burroni, D., Macchia, G.. Massone, A., Papini, E., Xiang Z., Figura, N. and Rappuoli, R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. U.S.A. 90 (1993) 5791-5795.
7. Covacci. A.. Tlliord, J. L., Giudice, G. D., Par- sonnet, J. and Rappuoli, R. Helicobacter pylori: virulence and genetic geography. Science 284 (1999)1328-1333.
8. DI: Wit, M. Y. L., Faber, W. R.. Krieg. S. R.. DouCiLas, J. T., Lucas. S. B., Montreewasuwat, N.. Pattyn, S. R.. Hussain, R., Ponnighaus, J. M.. Hartskllrl, R. A. and Klatser, P. R. Application of a polymerase chain reaction for the detection of Mycobacterium leprae in skin tissues. J. Clin. Microbiol. 29 (1991) 906-910.
9. De; Wit, M. Y. L. and Klatser. P. R. Mycobacterium leprae isolates from different sources have identical sequences of the spacer region between the 16S and 23S ribosomal RNA genes. Microbiology 140(1994) 1983-1987.
10. Dhandayuthapani, S.. Banu. M. J. and Kashiwabara, Y. Cloning and sequence determination of the gene coding for the elongation factor Tu of Mycobacterium leprae. J. Biochem. (Tokyo) 115 (1994) 664-669.
11. Doukhan, L., Predich. M.. Nair. G., Dussurget, O., Mandic-Mulec, I.. Coll. S. T., Smith. D. R. and Smith. I. Genomic organization of the mycobacterial sigma gene cluster. Gene 165 (1995) 67-70.
12. Draper, P. Cell walls of Mycobacterium leprae. Int. J. Lepr. 44 (1976) 95-98.
13. Fsihi, H. and Cole, S. T. The Mycobacterium leprae genome: systematic sequence analysis identifies key catabolic enzymes. ATP-dependent transport systems and a novel polA locus associated with genomic variability. Mol. Microbiol. 16 (1995) 909-919.
14. Gurtler, V. Typing of Clostridium difficile strains by PCR-amplification of variable length 16S-23S rDNA spacer regions. J. Gen. Microbiol. 139(1993)3089-3097.
15. Hunter, S. W. and Brlnnan, P. J. A novel phenolic glycolipid from Mycobacterium leprae possibly involved in immunogcnicity and pathogenicity. J. Bacteriol. 147 (1981) 728-735.
16. Imaeda.T., Kirchheimhr, W. F. and Barksdale, L. DNA isolated from Mycobacterium leprae: genome size, base ratio, and homology with other related bacteria as determined by optical DNA-DNA reassociation. J. Bacteriol. 150(1982)414-117.
17. Maeda, S., Ogura, K., Yoshida, H., Kanai, F., Ikenoue. T., Kato, N., Shiratori, Y. and Omuta. M. Major virulence factors, VacA and CagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut 42 (1998) 338-343.
18. Matar, C). M., Swaminathan, B., Hunter, S. B., Slater, L. N. and Welch, D. F. Polymerase chain reaction-based restriction fragment length polymorphism analysis of a fragment of the ribosomal operon from Rochalimaea species for subtyping. J. Clin. Microbiol. 31 (1993) 1730-1734.
19. McLaughlin, G. L., Howe, D. K.. Biggs, D. R., Smith, A. R., Ludwinski, P.. Fox, B. C., Tripathy, D. N., Frasch, C ,E., Wenger, J. D., Carey. R. B.. Hassan-King, M. and Vodkin, M. H. Amplification of rDNA loci to detect and type Neisseria meningitidis and other eubacteria. Mol. Cell. Probes 7 (1993) 7-17.
20. Park, M. H., Hwang, Y. S.. Park, K. S., Toku- naga, K.. Akaza, T., Juji, T. and Kim, S. I. HLA haplotypes in Koreans based oil 107 families. Tissue Antigens 51 (1998) 347-355.
21. Park, S. M, Park. J.. Kim, J. G., Clio. H. D., Clio, J. H., Lee. D. H, and Cha, Y. J. Infection with Helicobacter pylori expressing the cagA gene is not associated with an increased risk of developing peptic ulcer diseases in Korean patients. Scand. J. Gastroenterol. 33 (1998) 923-927.
22. Plikaytis, B. B., Gelber, R. H. and Shinnick. T. M. Rapid and sensitive detection of Mycobacterium leprae using a nested-primer gene amplification assay. J. Clin. Microbiol. 28 (1990) 1913-1917.
23. Pridich. M., Doukhan, L., Nair, G. and Smith, I. Characterization of RNA polymerase and two sigma-factor genes from Mycobacterium smegmatis. Mol. Microbiol. 15(1995) 355-366.
24. Shepard, C. C. and McRae. D. H. Hereditary characteristic that varies among isolates of Mycobacterium leprae. Infect. Immun. 3 (1971) 121-126.
25. van der Ende, A., Pan, Z.-J.. Bart, A., van der Hulst, R. W. M., Fellr, M., Xiao. S.-D., Tytgat, G. N. J. and Dankert, J. cagA-Positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect. Immun. 66 (1998) 1822-1826.
26. van sool1ngen, D., qlan, L., de haas, P. E. W., Douglas, J. T., Traore, H., Portaels, F., Qing, H. Z., Enkhsaikan, D., Nymadawa, P. and van Embden, J. D. A. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33 (1995) 3234-3238.
27. Williams, D. L., Gillis, T. P. and Portaels, F. Geographically distinct isolates of Mycobacterium leprae exhibit no genotypic diversity by restriction fragment length polymorphism analysis. Mol. Microbiol. 4 (1990) 1653-1659.
28. World Health Organization. Progress towards leprosy elimination. Wkly. Epidemiol. Rec. 73 (1998)153-160.
1. Ph.D.; Leprosy Research Center, National Institute of Infectious Diseases, 4-2-1 Aobacho, Higashimurayama-shi, Tokyo 189-0002, Japan.
2. M.D.
3. M.D., Ph.D., Institute of Hansen's Disease, College of Medicine, The Catholic University of Korea, 505 Banpo-Dong, Socho-ku, Seoul 137-701, Korea.
4. Ph.D., Molecular Biology Department, Laboratory Research Branch, GWL Hansen's Disease Center at Louisiana State University, P.O. Box 25072, Baton Rouge, LA 70894, U.S.A.
5. M.D., Ph.D., National Hospital Oshima Seishoen, 6034-1 Ajicho, Kidagun, Kagawa. Japan.
Present address for Dr. Kobayashi: Department of Microbiology, Osaka City University Medical School, 1-4-3 Asahi-machi, Abeno-ku, Osaka 545-8585, Japan.
Reprint requests to Dr. Kashiwabara at the above address or fax 81 -42-391-8212; e-mail: y-kashi@nih.go.jp
Received for publication on 28 March 2000.
Accepted for publication in revised form on 11 May 2000.