- Volume 66 , Number 1
- Page: 16–21
DNA amplification for detection of leprosy and assessment of efficacy of leprosy chemotherapy
ABSTRACT
Polymerase chain reaction (PCR) for the detection of Mycobacterium leprae was applied to fresh skin biopsies and slit-skin smears f rom 122 untreated leprosy patients. The PCR positivity rates in biopsies were 95.6% in multibacillary (MB) cases and 44.2% in paucibacillary (PB) cases. Following 1 month of treatment, MB cases declined by 54.3% and PB cases by 61.8% of initial values. Six-month values also declined f rom initial positivity rates to 50.3% and 53.8% of initial values in MB and PB, respectively. Larger declines in the rate of positivity were seen for skin-smear samples at 1 and 6 months in both MB and PB, but overall PCR positivity rates were lower than biopsy rates for M. leprae.RÉSUMÉ
On a utilisé une réaction de polymerase en chaîne (PCR) pour la détection de Mycobacterium leprae dans des biopsies cutanées fraîches et des frottis cutanés provenant de 122 malades de la lèpre non traités. Les taux de positivité de la PCR dans les biopsies étaient de 95,6%' dans les cas multibacillaires (MB) et 44.2% dans les cas paucibacillaires (PB). Après un mois de traitement, les taux de positivité diminuaient de 54,3% de leur valeur initiale chez les cas MB et de 61,8%- chez les cas PB. Les valeurs après six mois diminuaient aussi de respectivement 50,3% et 53,8% de leurs valeurs initiales chez les MB et les PB. Des diminutions plus importantes des taux de positivité étaient observées pour les frottis cutanés après un mois et six mois aussi bien chez, les MB que chez les PB, mais les taux globaux de positivité de la PCR pour M. leprae étaient plus faibles que les biopsies.RESUMEN
Se aplicó la reacción en cadena de la polimerasa (PCR) para la detección de Mycobacterium leprae en biopsias frescas de piel y en exudados de linfa cutánea de 122 pacientes con lepra sin tratamiento. La positividad de la PCR en las biopsias fue del 95.6% en la lepra multibacilar (MB) y de 44.2% en la lepra paucibacilar (PB). Después de 1 mes de tratamiento, la PCR declinó un 54.3% en los casos MB y un 61.8% en los casos PB. A los 6 meses de tratamiento, la positividad declinó al 50.3% en los casos MB y al 53.8% en los casos PB, en relación a los valores iniciales. En cuanto a los exudados de linfa cutánea, la caída en los porcentajes de positividad al mes y a los 6 meses de tratamiento fue más drástica tanto en los pacientes MB como en los PB, aunque desde el prinicipio la positividad global de la PCR fue más baja que en las biopsias de piel.The control of leprosy is based on case finding and chemotherapy. Early detection is important to prevent severe handicaps, spread of infection and economic loss. Considering the lack of an objective "gold standard" for the diagnosis of leprosy, clinical findings, accompanied by demonstration of leprosy bacilli in skin lesions through slitskin smears (14,24) in conjunction with histopathology (14,15), remain the only available standard for the diagnosis of leprosy. Serological assays (11) and skin tests (1), though simple, lack the required sensitivity and specificity to serve as diagnostic tools for Mycobacterium leprae infection. Although in vivo culture of M. leprae is another possibility (18), it is insensitive, labor-intensive and time-consuming.
Since the introduction of multidrug therapy (MDT), as advised by WHO (25), drugresistant M. leprae have been regularly reported. The current measurements of a patient's early response to chemotherapy, clinical observation and serial bacterial index (BI) and morphological index (MI) determination, are insensitive. The in vivo mouse foot pad technique is the only available tool for testing drug sensitivity; nevertheless it is too invasive, labor-intensive and time-consuming. Additional quantitative tests would be useful in assisting the clinician in evaluating the efficacy of the medication.
The recent development of the polymerase chain reaction (PCR) (7,26) has brought an unprecedented opportunity for sensitive, specific and rapid detection of leprosy bacilli in clinical specimens. In the study described here, we investigated the usefulness of PCR in comparison with more conventional methods (slit-skin smear and gelatin particle agglutination test) for the diagnosis of leprosy and the monitoring of leprosy patients during chemotherapy.
MATERIALS AND METHODS
Clinical specimens. Slit-skin smears and 4-mm punch, fresh skin biopsies taken for PCR were from newly diagnosed, untreated leprosy patients attending leprosy clinics in Khon Kaen, Samutprakarn and Bangkok, Thailand. Leprosy patients were diagnosed and classified clinically, bacteriologically and histopathologically according to the Ridley-Jopling scale (15,16) as lepromatous (LL), borderline lepromatous (BL), mid-borderline (BB), borderline tuberculoid (BT), tuberculoid (TT) or indeterminate (I). Multibacillary (MB) patients were defined as BT, BB, BL or LL patients having a BI of >0; whereas BT, TT or I patients with a BI of 0 were defined as paucibacillary (PB). The BI of each smear was determined by microscopic examination and the average BI was then calculated for each patient(14,24). Additional subsequent smear and biopsy materials were taken from the same lesions at 1 and 6 months after starting MDT; in this regimen PB cases receive supervised rifampin 600 mg once monthly and dapsone 100 mg every day for at least 6 months, MB cases receive supervised rifampin 600 mg once monthly, supervised clofazimine 300 mg once monthly, clofazimine 50 mg daily and dapsone 100 mg daily for at least 2 years.
Tissue biopsies from five individuals with dermatologic problems other than leprosy were also included as controls.
DNA extraction. The scalpel blade from a smear with tissue material on the tip was placed in a 1.5-ml sterile tube. Three hundred µl of lysis buffer containing 1 mg of proteinase K per ml and 0.05% Tween 20 in 100 mM Tris-HCl (pH 8.5) was added, the tube vortexed well, and then the blade was removed. The mixture was incubated at 60°C for 18 hr with mineral oil layered on top. Thereafter, the sample was incubated at 97°C for 15 min to inactivate the proteinase K.
The skin-biopsy specimen for PCR was minced with sharp scissors on a tissue grinder until lumps of tissue were no longer detected. To the tissue mince was then added 1 ml of sterile distilled water and the specimen was then thoroughly homogenized with a piston. DNA extraction by proteinase K digestion was performed as described above.
PCR. The specimens were subjected to PCR for the amplification of the 531-bp fragment of the pra gene encoding the species-specific 36-kDa antigen of M. leprae as described previously (7), with some modifications. Briefly, the 50-µl reaction volumes consisted of 10 mM Tris-HCl (pH 8.3), 50 mM KC1, 3.5 mM MgCl, 600 µM each dATP, dCTP, dGTP and dUTP, 200 ng each of primers S13 and S62 (7), and 1.25 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, Connecticut, U.S.A.). Twofold diluted samples with sterile distilled water were added to the mixtures. Sterile distilled water and DNA purified from M. leprae were included as negative and positive controls, respectively, in each experiment. M. leprae DNA was kindly supplied by M. J. Colston as part of the U.N. Development Programme/World Bank/World Health Organization Special Programme for Research and Training in Tropical Diseases. The amplification was then carried out in a thermocycler (Astec, Fukuoka, Japan) as follows: an initial denaturation step at 94°C for 10 min followed by 32 cycles consisting of denaturation at 94°C for 2 min, primer annealing at 60°C for 2 min and extension at 72°C for 3 min and then a final extension step at 72°C for 10 min. After amplification was finished, a 20-µl portion of the amplified product was run in a 1.5% (wt/vol) agarose gel in Trisborate EDTA (TBE) buffer at 70 V for 2 hr. After electrophoresis, the gel was stained with ethidium bromide, and the 531-bp DNA band was examined under an ultraviolet transilluminator.
Southern hybridization. For identification and confirmation of the amplified DNA in Southern blots (19), a sequence internal to the 531-bp M. leprae PCR fragment was amplified by using the nested primer set T3 and T4c, resulting in a 286-bp fragment which was then labeled with digoxigenin (Boehringer, Mannheim, Germany) (13). Details of the blot and hybridization were described elsewhere (23). Samples were scored positive when they were positive on the Southern hybridization.
Gelatin particle agglutination test. IgM antibody against phenolic glycolipid-I (PGL-I) in patient serum was detected by using Serodia-Leprae® (Fujirebio Inc., Tokyo, Japan), an M. leprae particle agglutination (MLPA) test, as described previously (8). A blood volume of 5 ml was collected from each patient before starting chemotherapy by venipuncture. The sera obtained were kept at -70°C until analysis with MLPA for the presence of anti-PGL-I antibody. The titer 1:32 was established as a cut-off value, defining a serum specimen which showed agglutination with less than 1:32 final dilution negative and that with equal to or more than 1:32 positive. If a pretreatment serum specimen was seropositive, then antibody titers of subsequent specimens at 1 and 6 months after starting chemotherapy would be measured.
Data analysis. All data were entered on a personal computer and analyzed using Epilnfo versions 5 and 6.
The PCR positivity rates in biopsies, smears and the MLPA seropositivity rate at the start of therapy were compared using ANOVA. The change of PCR, MLPA and BI values over time were analyzed by ANOVA, provided homogeneity of variance was fulfilled. If such was not fulfilled, then the Kruskal-Wallis ANOVA was used.
RESULTS
Specificity and sensitivity of PCR. The fact that the 531-bp DNA amplified with primers S13 and S62 is specific to M. leprae has been already well established (7). The sensitivity of the PCR protocol was determined by the presence or absence of the amplified 531-bp DNA band in Southern blot hybridization from a series of dilutions of purified M. leprae DNA template. PCR was shown to amplify DNA in amounts as low as 3.125 fg (approximately equivalent to 0.5 bacillus). The intensities of the 531bp DNA bands increased in a dose-response fashion as M. leprae DNA amounted equivalent to M. leprae increased in tenfold steps from 0.5 to 6.8 × 106 bacilli (result not shown).
A total of 122 untreated leprosy patients aged 5-76 years (average 38.5 years) were entered into this study, with the male:female ratio being 2.2. The patients were classified into 69 MB cases: 35 LL, 26 BL, 8 BT(+) and 53 PB cases: 45 BT(-), 5 TT, 3 I.
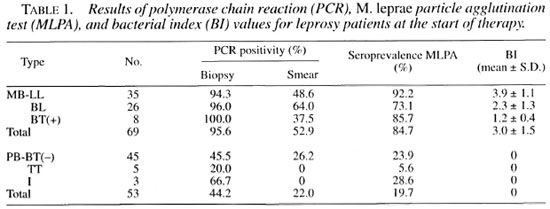
The results of PCR, MLPA and BI values at the start of therapy are shown in Table 1. Among the initial pretreatment specimens from MB patients, the PCR-biopsy positivity rate (95.6%) was significantly higher than those of PCR-smear (52.9%) and MLPA (84.7%) (p <0.001). In PB patients, the PCR-biopsy positivity rate (44.2%) was also significantly higher than those of PCR-smear (22.0%) and MLPA (19.7%) (p = 0.006).
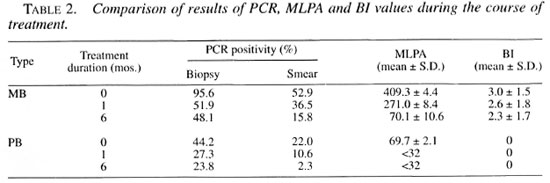
The PCR positivity rates, mean MLPA and mean Bl values + standard deviations during the course of treatment are presented in Table 2. After 1 month of therapy in MB and PB patients, all PCR positivity rates and mean MLPA were significantly lower (p <0.001) than the initial values at the beginning of therapy and stayed significantly lower at the 6-month period (p <0.001). No significant changes in the Bl values were noticed at 1- and 6-month periods (p = 0.052).
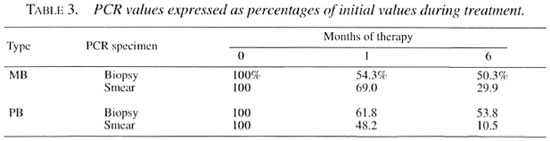
In order to allow comparison of PCR values during therapy, the PCR data were standardized by assigning the initial PCR positivity rate at the start of therapy to be 100%. Over the 6-month course subsequent values were calculated as a percentage of the initial value and they are presented in Table 3. The PCR percentages in biopsies declined to 54.3%-61.8% and those in smears declined to 48.2%-69.0% of the initial values after 1 month of therapy. By 6 months the PCR-smear percentages reflected an ongoing sharp decline to 10.5%-29.9% of the initial values; the PCR-biopsy values reflected a slower decline, being 50.3%53.8% of the original values.
Fresh skin biopsies from five patients with skin diseases other than leprosy gave negative results with PCR (data not shown).
DISCUSSION
Technical limitations in the diagnosis of leprosy can be overcome to a great extent by recent advances in the application of PCR. A number of PCR techniques targeting various genes of M. leprae have been developed recently (7,9,26). PCR has been found useful for a variety of specimens, including skin biopsy specimens and nasal swab specimens (5,6,9).
In this study, PCR applied on skin-biopsy specimens and skin-smear specimens was evaluated as a tool for diagnosing previously untreated leprosy patients compared to more conventional methods. The results (Table 1) showed that a relationship was found between disease severity and the values of PCR, MLPA and BI. PCR applied on biopsy specimens appeared to give positive results in 95.6% of MB patients and in 44.2% of PB patients, which were significantly higher than those of PCR applied on smear specimens and MLPA. The specificity of PCR has been confirmed by the absence of any false-positive results with specimens from controls. Because of high sensitivity and specificity, PCR - especially that applied on biopsy specimens - may be of value in confirmation of diagnosis in smear-negative, clinically/histologically atypical cases. There were two LL patients who gave repeatedly negative results with PCR applied on biopsy specimens while having rather low BI values of 2.0+ and 4.3+ and MLPA titers of 128 and 1024. respectively. These may result from uninformed previous antileprosy treatment.
PCR was also evaluated as a tool for monitoring leprosy patients under chemotherapy. At 1 and 6 months after starting therapy in MB patients, PCR positivity rates and the mean MLPA declined significantly, but no significant change in BI values was observed. The decline thus reflected the decrease in the load of live bacilli instead of the total bacterial load, as reported previously by Woods and Cole (26).
The declines of PCR positivity rates over time were analyzed by comparing the change in PCR percentages. The PCR percentages in biopsy specimens and smear specimens at 1 month of therapy declined to approximately 50% of initial values. Only the PCR percentages in smear specimens reflected a faster decline to approximately 10% - 30% of initial values at 6 months. The same trend was observed in PB patients whose AFB cannot be demonstrated and in mean MLPA, which was as low as 1:69.7 when beginning therapy, and which rapidly converted to seronegativity after 1 month of therapy. Thus, PCR applied on smear-positive specimens is more suitable as a tool for monitoring patients under chemotherapy.
The lower positivity rate in PCR applied on smear specimens compared to PCR biopsy may result from the small amount of tissue taken for DNA extraction and from contaminated blood as potential PCR inhibitor.
Nucleic acid sequence-based amplification (NASBA) (4) for the detection of 16S rRNA in skin biopsies indicated that NASBA may reflect the viability of M. leprae (20,21,22). However, application of NASBA in field work is limited at the moment because NASBA is still not easy to perform. Several indirect techniques for the determination of viability of M. leprae, such as fluorescent stainings (10,12), measurement of the ATP content (10), laser microprobe mass spectrometry (LAMMS) (17) and radiorespirometry (2,3), have been investigated and found limited in usage.
Although PCR is a very sensitive and specific method, it is quite expensive and sophisticated. Furthermore, the reproducibility of PCR needs to be verified before implementation. In areas where PCR facilities are accessible, PCR seems to be appropriate as a complementary tool in confirmation of the diagnosis of certain difficult or atypical cases. PCR applied on biopsy specimens is more suitable for diagnosis of leprosy patients than PCR applied on smear-positive specimens because of its higher sensitivity. However, PCR applied on smear specimens is worth trying because skin-smear technique is easy and practical in field work. PCR applied on smear specimens is more suitable for monitoring patients under chemotherapy because of the more rapid decline of the positivity rate during therapy that could be detected.
Acknowledgment. The authors wish to express their gratitude to Ms. S. Kamjan and Ms. S. Shuntawuttisettee for technical assistance and their thanks to the great efforts made by all field staff. This work was supported by Sasakawa Memorial Health Foundation.
REFERENCES
1. Bloom, B. R.,Convit, J., Godal., T., Noordeen, S. K., Perkins, F. T, Rees. R. J. W., Sansarricq, H., Shepard, C. C, Torrigiani, G. and Walter, J. Recommended safety requirements for the preparation of lepromin: a WHO Memorandum. Bull. WHO57(1979)921-923.
2. Chan, G. P., Garcia-Ignacio, B. Y., Chavez, V. E., Livelo, J. B., Jimenez, C. L., Parilla, M. L. and Franzblauau, S. G. Clinical trial of clarithromycin for lepromatous leprosy. Antimicrob. Agents Chemother. 38(1994)515-517.
3. Chan, G. P., Garcia-Ignacio, B. Y, Chavez, V. E., Livelo, J. B., Jimenez, C. L., Parilla, M. L. and Franzblau, S. G. Clinical trial of sparlloxacin for lepromatous leprosy. Antimicrob. Agents Chemother.38(1994)61-65.
4. Compton, J. Nucleic acid sequence-based amplification. Nature350(1991)91-92.
5. de Wit, M. Y. L., Douglas, J. T., McFadden, J. and Klatser, P. R. Polymerase chain reaction for detection of Mycobacterium leprae in nasal swab specimens. J. Clin. Microbiol.31(1993)502-506.
6. De Wit, M. Y. L., Faber, W. R., Krieg, S. R., Douglas, J. T., Lucas, S. B., Montreewasuwat, N., Pattyn, S. R., Hussain, R., Ponnighaus, J. M, Hartskeerl, R. A. and Klatser, P. R. Application of a polymerase chain reaction for the detection of M. leprae in skin (issue. J. Clin. Microbiol. 29(1991)906-910.
7. Hartskeerl, R. A., de Wit, M. Y. L. and Klatser, P. R. Polymerase chain reaction for the detection of Mycobacterium leprae. J. Gen. Microbiol. 135(19X9)2357-2364.
8. Izumi. S., Fujiwara, T., Ikeda, M., Nishimura, Y, Sugiyama, K. and Kawatsu, K. Novel gelatin particle agglutination test for serodiagnosis of leprosy in the field. J. Clin. Microbiol. 28(1990)525-529.
9. Jamil. S., Keer, J. T., Lucas, S. B., Dockrell, H. M., Chiang, T. J., Hussain, R. and Stoker, N. G. Use of polymerase chain reaction to assess efficacy of leprosy chemotherapy. Lancet 342(1993)264-268.
10. Katoch, V. M., Katoch, K., Ramanathan, U., Sharma. V. D., Shivannavar, C. T., Datta, A. K. and BHARADWAJ, V. P. Effect of chemotherapy on viability of Mycobacterium leprae as determined by ATP content, morphological index and FDA-EB fluorescent staining. Int. J. Lepr. 57(1988)615-621.
11. Klatser, P. R. Serology of leprosy. Trop Geogr. Med.46(1994)115-118.
12. Kvach, J. T. and Veras, J. R. A fluorescent staining procedure for determining the viability of mycobacterial cells. Int. J. Lepr. 50(1982)183-192.
13. Lion, T. and Hass, O. A. Nonradioactive labeling of probe with digoxigenin by polymerase chain reaction. Anal.Biochem. 188(1990)335-337.
14. Ridley, D. S. The bacteriological interpretations of skin smears and biopsies in leprosy. Trans. R. Soc. Trop. Med. Hyg.49(1955)449-452.
15. Ridley, D. S. Skin biopsy in leprosy. Basle: Documenta Geigy.1977.
16. Ridley, D. S. and Jopling, W. H. Classification of leprosy according to immunity - a five-group system. Int. J. Lepr. 34(1966)255-273.
17. Seydel, U., Haas, M., Rietschel. E. T. and Lindner, B. Laser microprobe mass spectrometry of individual bacterial organisms and of isolated bacterial compounds: a tool in microbiology. J. Microbiol. Meth.15(1992)167-183.
18. Shepard, C. C. A kinetic method for the study of activity of drugs against Mycobacterium leprae in mice. Int. J. Lepr. 35(1967)429-454.
19. Southern, E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98(1975)503-517.
20. van der Vliet, G. M. E., Cho, S. N, Kampirapap, K., Van Leeuwen, J., Schukkink, R. A. F., Van Gemen, B., Das, P. K., Faber, W. R., Walsh, G. P. and Klatser, P. R. Use of NASBA® RNA amplification for detection of Mycobacterium leprae in skin biopsies from untreated and treated leprosy patients. Int. J. Lepr. 64(1996)396-408.
21. van der Vliet, G. M. E., Schepers, P., Schukkink. R. A. F., van Gemen, B. and Klatser, P. R. Assessment of mycobacterial viability through RNA amplification. Antimicrob. Agents Chemother. 38(1994)1959-1965.
22. van der Vliet, G. M. E., Schukkink, R. A. F, van Gemen, B.,Schepers, P. and Klatser, P. R. Nucleic acid sequence-based amplification (NASBA) for the identification of mycobacteria. J. Gen. Microbiol. 139(1993)2423-2429.
23. Wichitwechkarn, J., Karnjan, S., Shuntawut-TISETTEE, S.,Sornprasit, C. Kampirapap, K. and PEERAPAKORN, S. Detection of Mycobacterium leprae infection by PCR. J. Clin. Microbiol. 33(1995)45-49.
24. Who Guidelines for Skin Smears. Int. J. Lepr. 55(1987)421-422.
25. Who Study Group. Chemotherapy of leprosy for control programmes. Geneva: World Health Organization. 1982. Tech. Rep. Ser. 675.
26. Woods, S. A. and Cole, S. T. A rapid method for the detection of potentially viable Mycobacterium leprae in human biopsies: a novel application of PCR. FEMS Microbiol. Lett.65(1989)305-310.
27. World Health Organization. Laboratory techniques for leprosy. Geneva: World Health Organization. 1987.
1. M.D.Leprosy Division, Sasakawa Research Building, Nonthaburi 11000, Thailand.
2. B.N., B.P.H.,Leprosy Division, Sasakawa Research Building, Nonthaburi 11000, Thailand.
3. Ph.D., N.H. Swellengrebel Laboratory of Tropical Hygiene, Royal Tropical Institute, 1105 AZ Amsterdam. The Netherlands.
4. B.Sc, Regional Office for Communicable Disease Control 6, Khon Kaen 40000, Thailand.
Reprint requests to Dr. Kowit Kampirapap, Phra-Pradaeng Hospital, Phra-Pradaeng, Samutprakarn 10130, Thailand.
Received for publication on 3 July 1997.
Accepted for publication in revised form on 7 January 1998.