- Volume 65 , Number 2
- Page: 190–6
Associations between HLA-DRB1 alleles and leprosy in an Indonesian population
ABSTRACT
To investigate whether the susceptibility to leprosy (type), subclinical infection with Mycobacterium leprae and the antibody response against M. leprae-specific antigens are associated with HLA-DR phenotypes sequence-specific oligonucleotide HLA-DRB1 and DQA1 typing and antibody assays have been performed in 79 leprosy patients (41 TT/BT and 38 LL/BL) and 50 healthy controls F rom a Javanese population in Yogyakarta, Indonesia. DRB 1*02 was associated with LL/BL [odds ratio (OR) 2.54, 95% confidence interval (CI) 0.97-9.78, p = 0.037 and attributable risk (AR) 41.5%] but not with TT/BT leprosy (p > 0.05). HLA-DRB1*12 was negatively associated with leprosy (either LL/BL or TT/BT [OR 0.33-0.35, p < 0.05, prevented fraction (PF) 58.8%65.3%]. No significant association was found between HLA-DRB1 or DQA1 type, anti-M. leprae antibody level and subclinical infection with M. leprae. These data indicate that in this population susceptibility to lepromatous leprosy is associated with HLA-DRB 1*02, while resistance to leprosy is associated with HLA-DRB 1*12. These associations are not paralleled with associations of the same HLA types with anti-M. leprae antibody level. Finally, the results of this study also support the notion that infection with M. leprae per se is not associated with HLA-DRB 1 or DQA1 alleles.RÉSUMÉ
Pour rechercher si la susceptibilité à la lèpre, l'infection sub-clinique pur le Mycobacterium leprae et la réponse d'anticorps vis-à-vis d'antigènes spécifiques de M. leprae sont associés à des phénotypes HLA-DR, on a réalisé le typage de séquences oligonucléotidiques spécifiques de HLA-DRB1 et DQA1 et des tests d'anticorps chez 79 malades de la lèpre (41 TT/BT et 38 LL/BL) et 50 témoins en bonne santé provenant d'une population javanaise de Yogyakarta, en Indonésie. DRB1 *02 était associé à LL/BL [odds ratio (OR) 2.54, limites de confiance (LC) à 95% 0.97-9.78, p = 0.037 et risque attribuable (RA) 41.5%], mais pas avec TT/BT (p > 0.05). HLA-DRB 1*12 était associé négativement avec la lèpre (soit LL/BL, soit TT/BT [OR 0.33-0.35, p < 0.05, fraction attribuable (FA) 58.8%65.3%]. Aucune association significative n'a été trouvée avec le type HLA-DRB 1 ou DQA1, le taux d'anticorps anti-M. leprae et l'infection subclinique par M. leprae. Ces données indiquent que dans cette population la susceptibilité à la lèpre lépromateuse est associée à HLA-DRB 1*02, tandis que la résistance à la lèpre est associée à HLA-DRB 1*12. Ces associations ne vont pas en parallèle avec des associations des mêmes types HLA avec le taux d'anticorps anti-M leprae. Finalement, les résultats de cette étude supportent également la notion que l'infection par M. leprae n'est pas associée par elle-même avec les alleles HLA-DRB 1 ou DQA1.RESUMEN
Para investigar si la susceptibilidad a la lepra, la infección subclínica, y la respuesta humoral contra los antígenos de Mycobacterium leprae están asociadas con determinados fenotipos HLA-DR, se analizaron las secuencias de oligonucleótidos específicas para HLADRB1 y DQA1 y la respuesta humoral contra antígenos de M. leprae en 79 pacientes con lepra (41 TT/BT y 38 LL/BL) y en 50 controles sanos de una población Javanesa en Yogyakarta, Indonesia. El antígeno DRB 1 *02 estuvo asociado con la lepra LL/BL [grado de predicción de 2.54, 95% de confianza interna (CI) 0.97-9.78, p = 0.037 y riesgo atribuible (RA) de 41.5%] pero no con la lepra TT/BT (p > 0.05). El antígeno HLA-DRB 1*12 se encontró negativamente asociado con la lepra en cualquiera de sus tipos [TP 0.33-0.35, p < 0.05, fracción evitada (FE) 58.8-65.3%]. No se encontró asociación significativa entre HLA-DRB 1 y el tipo de DQA 1, el nivel de anticuerpos anti-M. leprae, o la infección subclínica con M. leprae. Estos datos indican que en esta población, la susceptibilidad a la lepra lepromatosa está asociada con el antígeno HLA-DRB 1 *02, mientras que la resistencia a la enfermedad está asociada con HLA-DRB 1*12. Sin embargo, estas asociaciones no están relacionadas con las asociaciones de los mismos tipos HLA y el nivel de anticuerpos contra M. leprae. Finalmente, los resultados de este estudio también apoyan la noción de que la infección con M. leprae no está asociada per se con los alelos HLA-DRB 1 o HLADQA1.Leprosy is a chronic infectious disease of man caused by an intracellular microorganism, Mycobacterium leprae. Clinically, the disease is manifested as a spectrum with two polar forms, tuberculoid (TT) and lepromatous (LL). Between these two polar forms, there are intermediate forms: borderline tuberculoid (BT), midborderline (BB), and borderline lepromatous (BL). However, before clinical manifestations of the disease develop, patients may demonstrate a transitional form called indeterminate (I) which frequently heals spontaneously. The development of the clinical spectrum of the disease is related to the degree of cell-mediated immunity (CMI) and is influenced by genetic factors (4,5,8). Subclinical infection with M. leprae can be demonstrated by the presence of specific antibodies against M. leprae antigens in clinically healthy individuals (1,2,6,13).
Studies of genetics in leprosy have been reviewed by Fine (7,8). Among the genetic factors, human leukocyte antigens (HLA) have been studied extensively by several investigators (4,5,9,11,22-24). From these studies it became clear that HLA class II rather than HLA class I antigens are associated with disease susceptibility (22-23). Todd, et al. (22) on the basis of their own material in Louisiana and a meta-analysis of pooled reports of cases concluded that only HLA-DR2 and -DQ1 were associated with leprosy, in either tuberculoid or lepromatous forms. Based on his own and many other studies, de Vries (3) came to the conclusion that HLA-DR3 is often associated with TT leprosy and HLA-DQ1 universally with LL leprosy; whereas the susceptibility to M. leprae infection itself is not controlled by HLA-linked genes.
Leprosy still remains a public health problem in Indonesia. Although, the number of registered cases has decreased significantly since the implementation of multidrug therapy (MDT), the incidence of new cases is apparently unchanged (Hasibuan, Y. Situasi Penyakit Kusta di Indonesia dan Masalah-masalah Yang Dihadapi Dalam Pemberantasannya. Kumpulan Makalah Kongres Nasional VII Perdoski, Bukittinggi 9-12 Nopember 1992). The fundamental question of why some people contract leprosy (infection) and others do not still remains unanswered. One possibility is the genetic differences in susceptibility. In Indonesia, the distribution of leprosy is not uniform in either the prevalence or type of the disease. These differences may be due to the effect of many variables, including genetic factors. Since there has been no study thus far on the association between HLA and leprosy or M. leprae infection in Indonesia, we have conducted such a study in a Javanese population From Yogyakarta. We have used PCR-SSO DNA typing which is becoming more commonly used worldwide. Based on the earlier reports mentioned before, this study has focused on the HLA-DR and -DQ loci.
MATERIALS AND METHODS
Subjects. Seventy-nine leprosy patients (41 TT/BT, 38 LL/BL) and 50 healthy controls were recruited From Yogyakarta, Indonesia. They were all of the Indonesian/Javanese ethnic group. The diagnosis of leprosy was based on clinical, bacteriological and histopathological examinations, and patients were classified according to Ridley and Jopling (18). Blood samples were taken for HLA typing and serological assays and drawn before the start of chemotherapy for leprosy. The healthy controls consisted of 50 nonrelative household contacts (spouses). Another 50 healthy blood donors were used to determine the cut-off point for positivity in serological assays for the detection of anti-M leprae antibodies.
Serological assays. Sera collected From the subjects were tested for specific antibodies against M. leprae antigens, i.e., IgM anti-phenolic glycolipid-I (PGL-I) and IgG anti-36-kDa protein antigens, by using an ELISA and an Inhibition ELISA described previously (14,20). The semisynthetic DBSA antigen used in this study for the detection of anti-PGL-I antibodies has been standardized by the World Health Organization (WHO), and this antigen has demonstrated positive responses in the majority of leprosy patients. The 36-kDa protein antigen used in this study identified by Klatser, et al. (15) has not been studied extensivelyt hus far.
HLA-DRB1/DQA1 typing. For typing of HLA-DR/DQ alleles, we have used oligonucleotide hybridization essentially as previously described (10). DNA was extracted From peripheral blood lymphocytes. Amplification of this DNA was performed by the polymerase chain reaction (PCR) using sets of primers specific for DRB1 and DQA1, respectively. Typing for the alleles was carried out by dot blotting the PCR fragments and hybridization with synthetic oligonucleotides (10), allowing for low resolution DRB1 and for DQA1 high resolution typing (12). Using the available set of primers and SSO probes we could type for 12 HLA-DRB1 and 8 DQA1 specificities.
Statistical analysis. Frequencies of HLA-DRB1 and -DQA1 specificities in leprosy patients and controls were compared using a χ2-test (21). The strength of associations between the HLA specificities and (subclinical) leprosy was expressed as an odds ratio (OR) (17). Attributable risk (AR) or prevented fraction (PF) were calculated using the formula as suggested by Kleinbaum, et al. (16) and Kramer (17); p values were not corrected for multiple comparisions. These computations were performed using the SPSSPC version 3.1 and EPI-Info Statistical Software Packages.
RESULTS AND DISCUSSION
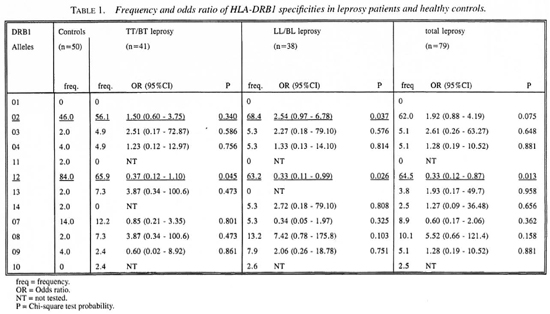
Table 1 presents the frequency distribution of the 12 HLA-DRB 1 specificities in cases and controls. DRB 1*01 was not found in this population. HLA-DRB 1*02 and DRB 1*12 showed significant associations with leprosy. For DRB 1 *02 a significant difference was found only between LL/BL leprosy and the healthy controls. This association had odds ratio (OR) of 2.54. The attributable risk (AR) was calculated to be 41.5%. A significant negative association was found between DRB 1*12 and both types of leprosy. The OR of this association was 0.33 for leprosy, 0.37 for TT/BT and 0.33 for LL/BL leprosy. Since the ORs were <1, we calculated the preventive fraction (PF) instead of the AR; these were 65.3%, 63.3% and 58.8%, respectively, for leprosy, LL/BL and TT/BT leprosy. To determine whether this association is secondary to the association with DRB 1 *02, an analysis was performed on DRB 1 *02 negative individuals. Also, among DRB 1 *02 negative individuals the frequency of DRB 1*12 was found to be higher in controls compared to both leprosy groups. This suggests that this association is a primary one, and that DRB 1*12 is a protective marker for leprosy in this Indonesian population.
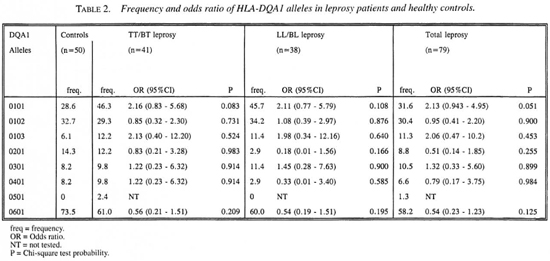
As shown in Table 2, eight DQA1* alleles have been identified. A high frequency was found for HLA-DQA 1*0601. HLA-DQA1 *0101 and DQA 1*0102 were distributed in moderate frequency; the remaining alleles were found in low frequency. No significant associations between DQA1* alleles and leprosy or leprosy subtypes were observed.
Next we analyzed whether the presence of certain HLA-DRB 1 or DQA alleles was correlated with the levels of IgM anti-PGL-I and IgG anti-M. leprae 36-kDa antibodies. This was separately analyzed for the patients and controls because patients had significantly higher antibody levels than did the controls. No significant differences in antibody levels were observed for the DRB 1*02 and DRB 1*12 specificities that were associated with leprosy or leprosy type (data not shown). So these data do not provide evidence for a possible immune response gene effect as the mechanism responsible for this/these association(s) (3-5). Some correlations were noted between several other DRB1 and DQA1 specificities and antibody levels but these were not significant after correction for the number of comparisons made.
Finally, we ascertained whether subclinical infection with M. leprae as determined by serology in household contacts was associated with HLA-DRB1 or DQA1 specificities. No association was noted between any of these HLA specificities and subclinical infection, neither based on the presence of IgM anti-PGL-I antibodies detected by ELISA (10 out of 50 contacts) nor IgG anti-36-kDa antibodies detected by Inhibition ELISA (15 out of 50 contacts). These numbers are too small to allow a strong conclusion, but they are at least in keeping with the notion that infection with M. leprae per se is not controlled by HLA-linked factors (3,4,5,7,19).
The results of this study demonstrate associations between DRB1 alleles and leprosy or leprosy type. DRB 1*02 shows a significant association with lepromatous (LL/BL) leprosy but not with tuberculoid (TT/BT) leprosy. This confirms previous studies in Japan (11) and India (19). In this study we have also found that HLA-DRB 1*12 has a negative association with leprosy. This is, indeed, a new finding that needs to be confirmed in future studies. If confirmed, the high frequency of DRB 1*12 in this population might be one factor which contributes to the low prevalence rate of leprosy in this population.
Acknowledgment. The authors would like to thank Zorana Grubic, Albert Naipal and Willem Verduyn for their help with the HLA typing. We also acknowledge the financial support of the Netherlands Leprosy Relief Association (NSL), the Commission of European Communities Directorate General for Science, Research and Development (grant TS2-0111-NL), and the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (Project Development Grant - ID. No. 880428).
REFERENCES
1. ABE, M., MANAGAWA, T, YOSHINO, Y, OZAWA, T, SAIKAWA, K. and SAITO, T. Fluorescent leprosy antibody absorption (FLA-ABS) test for detecting subclinical infection with Mycobacterium leprae. Int. J. Lepr. 48 (1980) 109-119.
2. CHANTEAU, S., CARTEL, J. L., Roux, J., PLICHART, R. and BACH, M.-A. Comparison of synthetic antigens for detecting antibodies to phenolic glycolipid-I in patients with leprosy and their household contacts. J Infect. Dis. 157 (1988) 770-776.
3. DE VRIES, R. R. P. HLA and disease: From epidemiology to immunotherapy. Eur. J. Clin. Invest. 22(1992)1-8.
4. DE VRIES, R. R. P., NIJENHUIS, L. E., LAI A FAT, R. F. M. and VAN ROOD, J. J. HLA-linked genetic control of host response to Mycobacterium leprae. Lancet 2 (1976) 1328-1330.
5. DE VRIES, R. R. P., OTTENHOFF, T. H. M. and SCHOOTEN, W. C. A. Human leucocyte antigen (HLA) and mycobacterial disease. Springer Semin. Immunopathol. 10 (1988) 305-318.
6. DOUGLAS, J. T. and WORTH, R. M. Field evaluation of an ELISA to detect antibody in leprosy patients and their contacts. Int. J. Lepr. 52 (1984) 26-33.
7. FINE, P. E. M. Immunogenetics of susceptibility to leprosy, tuberculosis and leishmaniasis; an epidemiological perspective. Int. J. Lepr. 49 (1981) 437-454.
8. FINE, P .E. M. Implications of genetics for the epidemiology and control of leprosy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 321 (1988) 365-376.
9. FINE, P. E. M., WOLF, E., PRITCHARD, J., WATSON, B., BRADLEY, D. J., FESTEINSTEIN, H. and CHACKO, C. J. G. HLA-linked genes and leprosy; a family study in Karagiri, South India. J. Infect. Dis. 140 (1979) 152-161.
10. GIPHART, M. J. and VERDUYN, W. The Eurotransplant HLA-DRB oligonucleotide typing set. Eur. J. Immunogenet. 18 (1991) 57-68.
11. IZUMI, S., SUGIYAMA, K., MATSUMOTO, Y. and OHKAWA, S. Analysis of the immunogenetic background of Japanese leprosy patients by the HLA system. Vox. Sang. 42 (1982) 243-247.
12. KIMURA, A. and SASAZUKI, T. Eleventh International Histocompatibility Workshop reference protocol for the HLA DNA-typing technique. In: HLA 1991. Tsuij, K., Aizawa, M. and Sasazuki, T, eds. Oxford: Oxford University Press, 1992.
13. KLATSER, P. R. Serology of leprosy Trop. Geogr. Med. 46 (1994) 59-62.
14. KLATSER, P. R., DE WIT, M. Y. L. and KOLK, A. H. J. An ELISA-inhibition test using monoclonal antibody for the serology of leprosy. Clin. Exp. Immunol. 56 (1985) 468-473.
15. KLATSER, P. R., VAN RENS, M. M. and EGGELTE, T. A. Immunochemical characterization of Mycobacterium leprae antigens of the SDS-polyacrylamide gel electrophoresis immunoperoxidase technique (SGIP) using patients' sera. Clin. Exp. Immunol. 56 (1984) 537-544.
16. KLEINBAUM, D. G., KUPPER, L. L. and MORGENSTERN, H. Epidemiologic Research. Principles and Quantitative Methods. Belmont, California: Lifetime Learning Publications, 1982.
17. KRAMER, M. S. Clinical Epidemiology and Biostatistics. A primer for Clinical Investigators and Decision Makers. Berlin: Springer-Verlag, 1988.
18. RIDLEY, D. S. and JOPLING, W. H. Classification of leprosy according to immunity; a five-group system. Int. J. Lepr. 34 (1966) 255-273.
19. SERJEANTSON, S. W. HLA and susceptibility to leprosy. Immunol. Rev. 70 (1983) 89-111.
20. SOEBONO, H. and KLATSER, P. R. A seroepidemiological study of leprosy in high- and low-endemic Indonesian villages. Int. J. Lepr. 59 (1991) 416-425.
21. SVEJGAARD, A., NIELSEN, L. S. and BODMER, W. F. HLA antigens and disease; statistical and genetical considerations. Tissue Antigens 4 (1974)95-105.
22. TODD, J. R., WEST, B. C. and McDONALD, J. C. Human leucocyte antigen and leprosy: study in northern Louisiana and review. Rev. Infect. Dis. 12(1990)63-74.
23. VAN EDEN, W. and DE VRIES, R. R. P. HLA and leprosy: a reevaluation. Lepr. Rev. 55 (1984)89-104.
24. YOUNGCHAIYUD U., CHANDANAYINGYONG, D. and VIBHATAVANIJA, T. The incidence of HLA antigens in leprosy. Vox Sang. 32 ( 1997) 342-345.
2. Ph.D.; Department of Immunohematology and Blood Bank, University Hospital, P. O. Box 9600, 2300 RC Leiden, The Netherlands.
3. Ph.D.; Department of Immunohematology and Blood Bank, University Hospital, P. O. Box 9600, 2300 RC Leiden, The Netherlands.
4. M.D., Department of Immunohematology and Blood Bank, University Hospital, P. O. Box 9600, 2300 RC Leiden, The Netherlands.
5. Ph.D., Department of Biomedical Research, Royal Tropical Institute, Meibergdreef 39, 1105 AZ Amsterdam, The Netherlands.
Reprint requests to Dr. R. R. P. de Vries.
Received for publication on 29 May 1996.
Accepted for publication in revised form on 29 January 1997.