- Volume 65 , Number 2
- Page: 197–202
No evidence for linkage between leprosy susceptibility and the human natural resistance-associated macrophage protein 1 (NRAMP1) gene in french Polynesia
ABSTRACT
In order to determine whether a human homolog (NRAMPl) to a murine candidate gene for resistance to mycobacteria influences susceptibility to human disease, we analyzed data F rom seven multicase leprosy families (84 individuals) F rom French Polynesia for linkage markers within the NRAMPl gene and leprosy per se. Individual family members were typed at nine polymorphic loci within NRAMPl. In addition, three physically linked, polymorphic microsatellite markers-D2S104, D2S173 and D2S1471-were also typed. Linkage analyses were done using affected sibpair and LOD score methods employing different modes of inheritance with full and reduced penetrance. The results of this study strongly suggest that NRAMPl is not linked to leprosy susceptibility in the French Polynesian families tested.RÉSUMÉ
Afin de déteminer si un homologue human (NRAMPl) d'un gène de souris candidat pour la résistance aux mycobactéries influence la susceptibilité a la maladie humaine, nous avons analysé les données de sept families comprenant des cas multiples de lèpre (84 personnes) en Polynésie française pour les marqueurs de lien dans le gène NRAMPl et la lèpre. On a typé les membres individuals des familles au niveau de neuf loci polymorphiques dans le NRAMPl. De plus, trois marqueurs microsatellites polymorphiques physiquement liés-D2S104, D2S173 et D2S1471 -ont également été typés. Des analyses de lien ont été effectuées par des méthodes de score de LOD sur des enfants de mêmes fratries atteints par la maladie, employant différents modes d'hérédité avec pénétration complète et réduite. Les résultats de cette étude suggèrent fortement que le NRAMPl n'est pas associé à la susceptibilité à la lèpre dans les familles de Polynésie française testées.RESUMEN
Para determinar si un gene humano (NRAMPl), homólogo a un gene murino de resistencia contra micobacterias, influye en la susceptibilidad a la lepra, analizamos los datos de 7 familias multicasos (84 individuos) de la Polinesia Francesa para buscar marcadores de ligamiento con el gene NRAMPl y con la lepra per se. Los miembros individuales de las familias correspondieron a tipos de nueve loci polimórlicos dentro de NRAMPl. Adicionalmente se tipificaron tres marcadores microsatélite polimórlicos fisicamente ligados (D2S104, D2S173 y D2S1471). Los análisis de ligamiento se hicieron usando el par afectado y métodos de registro LOD, considerando diferentes modos de herencia con penetrancias completa y reducida. Los resultados de este estudio sugieren fuertemente que NRAMPl no está ligado con la susceptibilidad a la lepra en las familias de la Polinesia Francesa estudiadas.Leprosy, caused by Mycobacterium leprae, is a chronic and debilitating disease that affects an estimated 5-6 million persons worldwide. There is now accumulating evidence that genetic factors play a significant role in human leprosy (1,5,12,14,16,17). The results of several segregation and pedigree analyses in large multigeneration families are consistent with a two-stage model for genetic control of susceptibility to leprosy, where susceptibility to disease per se is determined by autosomal genes (1,5,16) while HLA (human leukocyte antigen)linked genes control the specific clinical manifestation of the disease (12-14 ,7). One probable candidate for an autosomal human mycobacterial resistance gene is the homolog of the mouse Beg gene which controls murine resistance and susceptibility to infection with several species of mycobacteria including M. lepraemurium, M. bovis (BCG), and M. intracellular (13). We have previously investigated this hypothesis in leprosy families in French Polynesia by using genetic markers on the telomeric end of human chromosome 2q (9), which is syntenic with the proximal segment of the mouse Beg locus on chromosome 1 (13). Using these markers, no linkage was detected between leprosy susceptibility and the distal human chromosome 2 in French Polynesia.
Subsequent to that study, the Beg gene was cloned and shown to be identical with NRAMPI (18). Moreover, we cloned the human natural resistance-associated macrophage protein 1 (NRAMPI) gene on the chromosome 2q and identified nine sequence variants within the gene (3,10). Using these variants, we have recently detected significant linkage of leprosy susceptibility with NRAMPI in 16 Vietnamese multiplex leprosy families. These results prompted us to examine the role of NRAMPI for leprosy susceptibility among Polynesian leprosy families using these polymorphic markers within the NRAMPI gene.
MATERIALS AND METHODS
Patients and families. Seven multicase pedigrees with two (4 families) or three (3 families) generations and with at least one affected parent were identified in 1990. Families were identified From the records of the Institut Territorial Louis Malarde in Tahiti, French Polynesia. The entire study group of 84 individuals was composed of a total of 39 leprosy patients (24 males and 15 females). For more information about the patients and families see Levée, et al. (19).
Genotyping. Individual family members were typed at nine polymorphic loci previously mapped within NRAMPI (2,10). In addition three physically linked, polymorphic microsatellite markers, D2SI04, D2SI73 and D2S147I, were also typed. DNA samples used for typing were prepared as described by Levée, et al. (9).
Polymerase chain reaction (PCR) amplifications were carried out in 30-µl reaction volume containing 100 ng of genomic DNA, 50 mM Tris-HCl (pH 8.3), 1.8 mM MgCl2, 0.05% Tween-20, 0.05% NP40, 250 µM of each dNTP, 0.5 µM of each primer and 2 U of Taq polymerase. The primers used for amplification are described elsewhere (10).
Samples were processed through one step of denaturation (94ºC for 3 min) followed by 10 cycles of a touch-down program (30 sec at 94ºC, 1 min at 66ºC with -0.5ºC/cycle increments and 1 min at 72ºC). The touch-down phase was followed by 25 cycles of denaturation (94ºC for 30 sec), annealing (55ºC for 1 min), elongation (72ºC for 1 min), and one last step of elongation at 72ºC for 7 min. The forward primers used to amplify the DNA segments carrying the sequence variants in the region 3'UTR, 5'UTR and the microsatellite markers D2S104, D2S173, D2S1471 were labeled with γ-32P-ATP (Amersham Corp., Arlington Heights, Illinois, U.S.A.) and the variants were detected on 6% polyacrylamide denaturing gels run at room temperature at constant power of 70 watts for 2-3 hr. For the other sequence variants, restriction-endonuclease digestions were done on the amplified DNA products by using 5 U of enzyme per PCR reaction under conditions recommended by the supplier (New England Biolabs, Beverly, Massachusetts, U.S.A.). Restriction-enzyme digestion products were resolved by electrophoresis on 12% polyacrylamide gels and stained with ethidium-bromide for visualization on standard ultraviolet (UV) boxes. All genotypes were interpreted by two independent observers.
Linkage analyses. Linkage analyses were carried out using sibpair and LOD score methods. For the analyses, susceptibility to leprosy per se (any form of leprosy) was used as the phenotype to be analyzed.
The affected sibpair method described by de Vries, et al. (4) was used to detect the segregation of parental haplotypes in sibships with at least two affected siblings. The observed difference (F) between the number of affected siblings with one and those with the other parental haplotype was compared with the expected difference (f) for random segregation using the statistic test: (|Σ F- Σfl - 0.5)7 Σv2 (v2 is the variance of f and Σf and ΣF are the summations of f and F values over all sibships). This test has a chi-squared distribution of 1 df. The employed method has the advantage of accounting in a natural way for families with more than two affecteds and allows the analysis of families where only one parent is informative for the analysis of allele sharing. The extended haplotypes comprising NRAMP1 and microsatellite markers were tested. For this analysis family 4 was divided in three nuclear families. Families 1, 2, 6 and 7 were excluded From the analysis because families 1 and 2 were not informative and families 6 and 7 had only one affected sibling per generation.
LOD score analyses were carried out using the LINKAGE 5.1 computer program package (8). Two point LOD scores were calculated with the MLINK program using NRAMP1 haplotypes. LOD scores obtained at different recombination fractions were added over all families. Under the assumption of Hardy-Weinberg equilibrium, the disease gene frequency was defined for each mode of inheritance: 0.0471 for recessive, 0.0011 for dominant and 0.0022 for codominant mode of inheritance following the reasoning given by Levee, et al. (9). Successive linkage analyses were performed for the three modes of inheritance assuming penetrances of 100% and 50% of the phenotype as well as with affecteds only.
RESULTS
We defined the NRAMP1 allele frequencies in the French Polynesian population by genotyping 20 unrelated individuals who are part of the seven analyzed families for nine intragenic NRAMP1 sequence variants. The comparison of NRAMP1 allele frequencies among Caucasian, Asian and Polynesian is summarized in Table 1. Most tested variants show a significant lower heterozygosity in Polynesians as compared to Asian and Caucasian individuals.
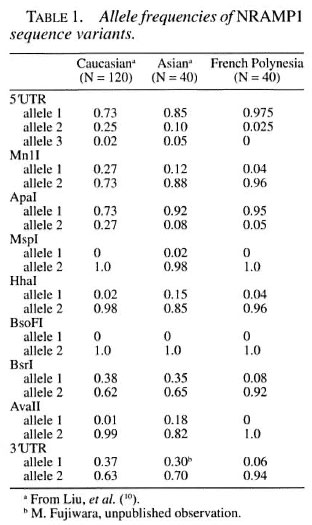
Since available NRAMP1 polymorphisms showed very limited informational content for the French Polynesian families, we expanded the marker panel by three additional microsatellite markers (D251471, D2S173, D2S104) which have been shown to be located within 1 Mbp of NRAMP1 (10). Haplotypes overlapping the NRAMP1 locus were constructed From familial segregation patterns, and all of the parental chromosomes (except in family 1 and 2) were found informative for analysis of allelic segregation.
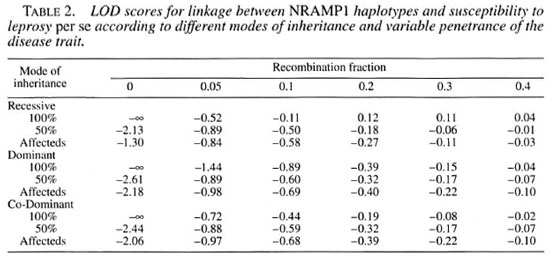
We first tested for possible linkage between the NRAMP1 haplotype and leprosy susceptibility using the LOD score method. Estimates for disease allele frequencies were taken From Levee, et al. (9). Since no further information was available with respect to the possible genetic model of leprosy susceptibility, we tested three modes of inheritance of the putative susceptibility gene with varying degrees of penetrance of the susceptibility phenotype (Table 2). Regardless of the mode of inheritance and the level of penetrance assumed, no evidence for linkage of leprosy susceptibility with NRAMP1 was obtained. Conversely, linkage was excluded for most models tested, and in cases where exclusion was not statistically significant this appears entirely due to the lack of power contained in the family structure rather than to a possible effect of NRAM PI on the computed LOD scores.
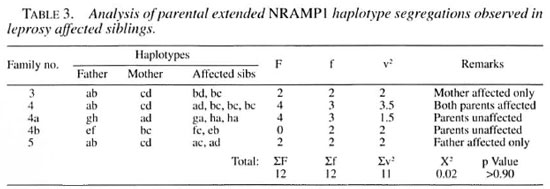
To avoid pitfalls inherent to incomplete knowledge of the genetic parameters we also employed nonparametric affected sibpairs analysis. The probability of random parental haplotype segregation into affected sibpairs was tested with the analytical procedure of de Vries, et al. (4). The results of this analysis are summarized in Table 3, and show no significant deviation From random segregation of NRAMP1 into leprosyaffected offspring. This result argues strongly against a role of NRAMP1 in the control of leprosy susceptibility among the French Polynesians leprosy families tested.
DISCUSSION
Prior to the cloning of the human NRAMP1 gene and the availability of its polymorphic markers, we employed markers on the distal human chromosome 2 in order to test for linkage between leprosy susceptibility in French Polynesia and the putative human homolog of the Beg gene (9). In addition, Shaw and co-workers employed the same approach in family studies conducted in Pakistan and Brazil (15). In both of these studies, no linkage was detected between chromosome 2q markers and susceptibility to leprosy. In the present study, we have used nine sequence variants within NRAMPI gene, which we have identified subsequent to the cloning of human NRAMPI in 1994 (3,10) in order to confirm our original findings in French Polynesia. The results of the new linkage analysis which directly tested for linkage of NRAMPI gene with leprosy confirmed that this gene is not linked with leprosy susceptibility in this population. Moreover, the present results confirm our earlier results using chromosome 2q markers and represent the validity of the close-marker associated approach in other situations where the chromosomal location of a candidate locus is known before direct markers for the gene in question are available.
The results of our present study also provide additional information on the genetic distribution of the French Polynesian population. In comparison with Caucasians and Asians, the markers within NRAMPI and the microsatellite markers tested in this study showed lower heterozygosities in French Polynesians. These results are in agreement with other studies showing that Polynesians have lower heterozygosities at minisatellite loci than other populations in Oceania (6,7,11). The genetic homogeneity in the French Polynesian population could be the result of the Polynesians having passed through one or several population bottlenecks and/or a small population size has persisted for many generations. The low heterozygosity of the Polynesians makes them a unique population for genetic studies since one would expect to detect a strong effect of the gene if the susceptibility allele would have been selected in this population. Thus we can speculate that the NRAMPI gene has little or no role in leprosy susceptibility in French Polynesia because the susceptibility allele did not segregate in this population.
Finally, using these same NRAMPI variants, we have recently detected significant linkage of leprosy susceptibility with NRAMPI in 16 Vietnamese multiplex leprosy families (submitted). This observation is supported by our recent segregation analysis showing that Mendelian inheritance of leprosy susceptibility could be detected in Vietnamese but not Chinese families From the same endemic area (1). The results of the present study showing that leprosy susceptibility is not linked to NRAMPI in Polynesian families offer further support that susceptibility to leprosy is a genetically heterogeneous trait. The reason why an effect of NRAMPI alleles is observed in only some populations is unknown. At present, the most parsimonious explanation is that leprosy susceptibility is under multigenic control, and that the effect of NRAMPI is more pronounced on a specific but hitherto unknown genetic background. Alternatively, it is possible that environmental and/or microbial components differ sufficiently among different leprosyaffected populations to either mask or enhance the action of specific susceptibility loci. These and similar hypotheses need to be tested with a larger data set of affected pedigree pairs for leprosy in different ethnic populations.
Acknowledgment. The authors wish to thank Dr. E. Buschman for help in writing the paper. M. Roger is the recipient of a fellowship From the McLauglin Foundation Canada. E. Schurr is a chercheur-boursier of the Fonds de la Recherche en Santé du Québec (FRSQ). This work was supported by a grant From the Canadian Genome Analysis and Technology Program (CGAT) and From Association Raoul Follereau.
REFERENCES
1. ABEL, L., LAP, V. D., OBERTI, J., THUC, N.V., CUA, V. V., GUILLOUD-BATAILLE, M., SCHURR, E. and LAGRANGE, PH. Complex segregation analysis of leprosy in Southern Vietnam. Genet. Epidemiol. 12 (1995)63-82.
2. BUU, N. T, CELLIER, M., GROS, P. and SCHURR, E. Identification of highly polymorphic length variant in the 3'UTR of NRAMPl. Immunogenetics 42 (1995)428-429.
3. CELLIER, M., GOVONI, G.. VIDAL, S. M., KWAN, T, GKOULX, N., LIU, J., SANCHEZ, F., SKAMENE, E., SCHURR, E. and GROS, P. Human natural resistance-associated macrophage protein: c-DNA cloning, chromosomal mapping, genomic organization and tissue-specific expression. J. Exp. Med. 180(1994)1741-1752.
4. DE VRIES, R. R. P., NIJENHUIS, L. E., LAI A FAT, R. F. M. and VAN ROOD, J. J. HLA-linked genetic control of host response to Mycobacterium leprae. Lancet 2 (1976) 1328-1330.
5. FEITOSA, M. F., BORECKI, I., KREIGER, H., BEIGUELMAN, B. and RAO, D. C The genetic epidemiology of leprosy in Brazilian population. Am. J. Hum. Genet. 56 (1995) 1179-1185.
6. FLINT, J., BOYCE, A. J., MARTINSON, J. J. and CLEGG, J. B. Population bottlenecks in Polynesia revealed by minisatellites. Hum. Genet. 83 (1989) 257-263.
7. HERTZBERG, M. S., MICKLESON, K. N. P. and TRENT, R. J. Limited genetic diversity in Polynesians reflected in the highly polymorphic 3'HVR-globin gene marker. Hum. Hered. 42 (1992) 157-161.
8. LATHROP, G. M., LALOUEL, J. M., JULIER, C. and OTT, J. Strategies for multilocus linkage analysis in humans. Proc. Natl. Acad. Sci, U.S.A. 81 (1984) 3443-3446.
9. LEVEE, G., LIU, J., GICQUEL, B., CHANTEAU, S. and SCHURR, E. Genetic control of susceptibility to leprosy in French Polynesia; no evidence for link age with markers on the telomerie human chromosome 2. Int. J. Lepr. 62 (1994) 499-511.
10. LIU, J., FUJIWAKA, T. M., BUU, N. T., SANCHEZ, F. O., CELLIER, M., PARADIS, A. J., FRAPPIER, D., SKAMENE, E., GKOS, P., MORGAN, K. and SCTIURR, E. Identification of polymorphisms and sequence variants in the human homologue of the mouse natural resistance-associated macrophage protein gene. Am. J. Hum. Genet. 56 (1995) 845-853.
11. MARTINSON, J. J., HARDING, R. M., PHILIPPON, G., FLYE SAINTE-MARIE, F., ROUX, J., BOYCI;, A. J. and CLEGG, J. B. Demographic reductions and genetic bottlenecks in humans: minisatellite allele distributions in Oceania. Hum. Genet. 91 (1993 ) 445-450.
12. OTTENHOEE, T. H. and DE VRIES, R. R. P. HLA class II immune response and suppression genes in leprosy. Int. J. Lepr. 55 (1987) 521-534.
13. SCHURR, E., BUSCHMAN, E., MALO, D., GROS, P. and SKAMENE, E. Immunogenetics of mycobacterial infections: mouse-human homologies. J. Infect. Dis. 161 (1990) 634-641.
14. SERJEANTSON, S. HLA and susceptibility to leprosy. Immunol. Rev. 70 (1983) 89-112.
15. SHAW, A. M. ATKINSON, S., DOCKRELL, H., HUS-SAIN, R., LINS-LAINSON, Z., SHAW, J., RAMOS, F., SII.VEIRA, F, MEIIDI, S. Q., KAUKAD, F., KHALIQ, S., CHIANG, T. and BLACKWELL, J. An RFLP map for 2q33-q37 From multicase mycobacterial and leishmanial disease families: no evidence for an Lsh/lty/Bcg gene homologue influencing susceptibility to leprosy. Ann. Hum. Genet. 57 (1993) 251-271.
16. SHIELDS, E. D., RUSSELL, D. A. and PERICAK-VANCE, M. A. Genetic epidemiology of the susceptibility to leprosy. J. Clin. Invest. 79 (1987) 1139-1143.
17. VAN EDEN, W. and DE VRIES, R. R. P. HLA and leprosy: a réévaluation. Lepr. Rev. 55 (1984) 89-104.
18. VIDAL, S. M., MALO, D., VOGAN, K., SKAMENE, E. and GROS, P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Beg. Cell 73 ( 1993) 469-485.
1. M.D., Ph.D.; McGill Center for the Study of Host Resistance, Montreal General Hospital Research Institute, Room LI 1-521. 1650 Cedar Avenue, Montreal, Quebec, Canada H3G 1 A4.
2. Ph.D., McGill Center for the Study of Host Resistance, Montreal General Hospital Research Institute, Room LI 1-521. 1650 Cedar Avenue, Montreal, Quebec, Canada H3G 1 A4.
3. Ph.D.; McGill Center for the Study of Host Resistance, Montreal General Hospital Research Institute, Room LI 1-521. 1650 Cedar Avenue, Montreal, Quebec, Canada H3G 1 A4.
4. Ph.D., Institut Territorial de Recherches Médicales Louis Malarde, B. P. 30, Papeete, Tahiti, Polynésie Française.
5. Ph.D., Unite de Génétique Mycobacterienne, Institut Pasteur, 25 Rue du Dr. Roux, 75274 Paris 15, France.
Reprint requests to Dr. Schurr at the address above or FAX 514-933-7146.
Received for publication on 1 October 1996.
Accepted for publication in revised form on 6 March 1997.