- Volume 71 , Number 2
- Page: 115–8
Leprosy susceptibility revealed1
ABSTRACT
In order for these findings to have practical significance in terms of leprosy control and prevention, it will be necessary to extend the linkage of chromosome 6q25 to another region endemic for leprosy. Replicative findings would likely mean that the chromosome 6q25 susceptibility gene is a variant of a common gene that promotes susceptibility to infection per se. Identifition of the gene variant will hopefully reveal insight about transmission and disease incidence-the longstanding enigmas of leprosy. Whether a more effective, universal MDT treatment or another type of prevention (either vaccine or environmental) could be based on this knowledge, is an exciting prospect to contemplate.Although global leprosy rates have declined since the introduction of multidrug therapy (MDT) in the 1980's, in certain countries the disease incidence remains surprisingly high (19). Over 700,000 new cases were reported last year, mostly in India, Brazil, and South East Asia (19). The reasons for the continued transmission of leprosy, in an era of extensive MDT coverage, are not understood. Humans are the reservoir for Mycobacterium leprae, the causative agent of leprosy, a disease well documented to be transmitted by close contact. The fact that only a minority of people exposed to the bacillus develop clinical leprosy suggests that underlying factors greatly influence susceptibility. A substantial component of leprosy susceptibility is inherited, as shown by complex segregation analyses of leprosy and the high concordance rates of leprosy among monozygotic twins (4,7,17). The identification of genes with major effects would be useful knowledge in the strategy of leprosy control and elimination. Until recently, however, limitations in dissecting the complex (i.e., non-Mendelian) genetics of susceptibility to infectious diseases such as leprosy have restricted studies to analyses of individual variants of known genes with small effect on the risk of developing leprosy.
BACKGROUND
The current work of Mira, et al. (13) [see Current Literature, p. 181] has been brought about by developments in the field of leprosy and in the field of genomics. Since the early 1980's, studies have documented that different genes determine susceptibility to leprosy per se, and the clinical subtypes of paucibacillary (PB) and multibacillary (MB) leprosy (2,8,9). These studies detected a major effect of non-HLA genes on leprosy per se, whereas PB and MB leprosy were found to be partly controlled by HLA genes. It was also recognized that the two-stage pattern of genetic control is reminiscent of mouse mycobacterial infection, where innate, non H-2 genes control early replication of bacteria (i.e., the Nramp1 gene) and H-2 genes partly control the acquired immunity to infection (6). In searching for the effect of major genes controlling susceptibility to leprosy, several genes were found to have small effects primarily on leprosy subtypes, such as the NRAMP1, TNFA and VDR genes, but again, any major gene effects remained unaccounted for. The human genome project has now provided geneticists with the ability to perform 'whole genome analysis.' This approach has the advantage that the entire genome can be scanned in an unbiased fashion for the presence of susceptibility genes. A study in 2001 (18) conducted the first whole genome analysis on PB leprosy in India, and found a locus on chromosome 10p13 linked with the PB subtype. The recent Mira, et al. paper has made another step forward as the first study to combine a full analysis of leprosy per se, at the whole genome level.
THE RESEARCH TEAM
The Mira, et al. study team was comprised of four groups spread across three continents. The Vietnamese families participated with the aid of an excellent clinical infrastructure and leprosy control at the Hospital for Dermato-Venereology in Ho Chi Minh City. The leprosy epidemiology team has a stringent ethic of pedigree collection and a precise clinical analysis of leprosy and its subtypes. This is particularly crucial as PB cases are not as easily discerned from other disease conditions as MB leprosy. The statistical analysis group, at the INSERM U550, Necker Medical School in Paris, has been working on genetic analysis of infectious disease for many years. In fact, the group provided crucial support for the two stage genetic model for leprosy based on a study of leprosy inheritance in Desirade Island (2). Recently this group developed the "maximum likelihood binomial" (MLB) method for linkage analysis in families with high numbers of affected children and used in the Mira, et al. paper (1). In Montreal, the McGill Centre for the Study of Host Resistance is home to the discovery of the Bcg/Nramp locus, and researchers at the Centre have studied genetic susceptibility to mycobacteria in humans for many years. In addition, the Genome Quebec Innovation Centre where the large scale mapping was carried out, is a leading center for genomic technologies.
RESULTS
In Vietnam, it had been previously found that leprosy per se was linked to a major gene (4) and so a whole genome analysis was undertaken to locate the gene(s). The 86 multiplex families (i.e., families with multiple affected children) in the study comprised over 200 leprosy affected children that presented pauci- and multibacillary forms of leprosy in approximately equal numbers. Genomic DNA was obtained from all affected children and their parents, and each DNA was genotyped with 388 anonymous microsatellite markers covering the whole genome at approximately 10 centimorgan (cM)* intervals. The microsatellites employed are highly polymorphic repeat length markers that "tag" all four parental chromosomes. Hence, the inheritance of specific parental chromosomal regions to affected children can be followed with high accuracy. If a chromosomal region has no linkage to a gene predisposing to leprosy, it is expected that over a large number of families pairs of affected siblings (representing 4 parental chromosomes) will share one copy of the same chromosomal region in 50% of all cases, i.e., the same chromosome from one parent was inherited by both sibs while two different chromosomes were inherited by the two sibs from the other parent (The Figure ). In addition, 25% of all sib pairs will either share no chromosomal region or both. Any significant excess of sharing of chromosomal regions among affected siblings is indicative for the presence of a leprosy susceptibility gene on the shared chromosomal segment. The MLB method used by Mira, et al. expands this analysis to families with more than 2 affected siblings. Analysis of sharing of microsatellite alleles showed that 11 chromosomal regions were possibly linked to leprosy per se. Next, to better reveal true linkages, the chromosomal segments underlying the 11 linkage peaks were genotyped for an additional 89 markers spanning the target regions at approximately 1 cM intervals. The results obtained with this so-called high resolution linkage mapping demonstrated that only one region, 6q25 on the long arm of chromosome 6, was significantly linked to leprosy susceptibility. Criteria for significant linkage of a chromosomal region to a disease phenotype are very stringent and nowadays, only odds of approximately 10,000: 1 are considered significant. It is customary to express the odds in favour of linkage as a logarithm to base 10, so called "lod" scores. The lod score for linkage of chromosome region 6q25 to leprosy was found to be 4.31. In addition, a lod score of 2.62 was obtained for linkage of leprosy susceptibility to the HLA region on the short arm of chromosome 6. The latter, while not significant, is considered "suggestive" of linkage and it seems likely that HLA loci influence leprosy susceptibility, as previously suggested by other studies (16).
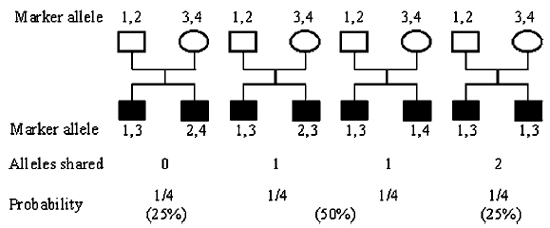
The figure. Expected allele sharing among affected sibpairs if marker and disease are unlinked.
Based on the excess of shared alleles, Mira, et al. then computed the risk to develop leprosy of siblings of affected patients relative to the general population as 2.21. This so called s value needs to be distinguished from the more commonly used relative risk measure in cohort studies. Indeed, the relationship between these two risk measures is not straightforward, and it is difficult to predict into what relative risk values the observed s value of 2.21 will transform (10).
Linkage analysis of complex traits such as susceptibility to leprosy is a tricky game and the possibility of false positive results is always present despite the stringent criteria employed. To counter this possibility, Mira, et al. enrolled a second independent sample of 208 simplex families (i.e., families with one affected child). Simplex families are of little use for linkage studies but are exquisitely suited to test for association between genetic markers and phenotype. Mira, et al. then employed 7 microsatellite markers directly underlying the major linkage peak detected in the 86 multiplex families and tested individual microsatellite alleles for association with leprosy susceptibility. Since microsatellites are very polymorphic, i.e., contain many alleles, this approach has the major drawback that many tests need to be conducted, strongly increasing the possibility that a detected association may be due to chance. To adjust for this scenario, Mira, et al. made two critical corrections, first considering for association analysis only those alleles that were informative in at least 20 families, and, second, all p-values obtained for individual alleles were multiplied by the total number of alleles tested. Even under these stringent conditions, two alleles at markers D6S1035 and D6S305 were found highly significantly associated with leprosy susceptibility providing confirmation of a leprosy locus on chromosome 6q25 independent of the linkage results in the multiplex families.
The Mira, et al. study also has clarified results obtained in the Indian study (18) that detected a locus for susceptibility to leprosy on human chromosome 10p13. Since the Indian families comprised only paucibacillary cases, it was not clear if this locus was a leprosy susceptibility gene per se, or a susceptibility gene for paucibacillary leprosy only. Mira, et al. were able to demonstrate, by testing linkage of leprosy per se and paucibacillary leprosy independently in their families, that the locus on chromosome 10p13 was indeed a susceptibility gene for paucibacillary leprosy only. By contrast, the newly detected chromosome 6q25 locus was linked to susceptibility to both pauci- and multibacillary forms of leprosy. However, if the locus on chromosome 6q25 is a susceptibility locus for leprosy per se, why was this locus not detected in the Indian families? There are many possible answers to this question, but the definite answer will only emerge once the underlying molecular defect predisposing individuals to higher risks of leprosy susceptibility per se in the Vietnamese has been identified. Testing of such risk variants in the Indian population will give the answer if the same genetic mechanisms are operating in leprosy patients of different ethnic and racial backgrounds.
- Ellen Buschman, Ph.D. and Emil Skamene, M.D., Ph.D.
McGill Centre for the Study of Host Resistance
1650 Cedar Avenue
Montreal, PQ, H3G 1A4, Canada
REFERENCES
1. Abel, L., Alcais, A. and Mallet, A. Comparison of four sib-pair linkage methods for analyzing sibships with more than two affecteds: interest of the binomial maximum likelihood approach. Genet. Epidemiol. 15(1998)371-390.
2. Abel, L. and Demenais, F. Detection of major genes for susceptibility to leprosy and its subtypes in a Caribbean island: Desirade island. Am. J. Hum. Genet. 42(1988)256-266.
3. Abel, L., Sanchez, F.O., Oberti, J., Thuc, N.V., Hoa, L.V., Lap, V.D., Skamene, E., Lagrange, P.H. and Schurr, E. Susceptibility to leprosy is linked to the human NRAMP1 gene. J. Infect. Dis. 177(1998)133-145.
4. Abel, L., Vu, D.L., Oberti, J., Nguyen, V.T., Van, V.C., Guilloud-Bataille, M., Schurr, E. and Lagrange, P.H. Complex segregation analysis of leprosy in southern Vietnam. Genet. Epidemiol. 12(1995)63-82.
5. Alcais, A., Sanchez, F.O., Thuc, N.V., Lap, V.D., Oberti, J., Lagrange, P.H., Schurr, E. and Abel, L. Granulomatous reaction to intradermal injection of lepromin(Mitsuda reaction)is linked to the human NRAMP1 gene in Vietnamese leprosy sibships. J Infect Dis. 181(2000)302-308.
6. Casanova, J.L. and Abel, L. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 20(2002)581-620.
7. Chakravarti, M.R. and Vogel, F. A twin study on leprosy. In: Topics in Human Genetics. New York: Georg Thieme Verlag Stuttgart, 1973, pp. 1-123.
8. van Eden, W., Gonzalez, N.M., de Vries, R.R., Convit, J. and van Rood, J.J. HLA-linked control of predisposition to lepromatous leprosy. J. Infect. Dis. 151(1985)9-14.
9. van Eden, W., de Vries, R.R., Mehra, N.K., Vaidya, M.C., D'Amaro, J. and van Rood, J.J. HLA segregation of tuberculoid leprosy: confirmation of the DR2 marker. J. Infect. Dis. 141(1980)693-701.
10. Goldgar, D.E. Population aspects of cancer genetics. Biochimie. 84(2002)19-25.
11. Meisner, S.J., Mucklow, S., Warner, G., Sow, S.O., Lienhardt, C. and Hill, A.V. Association of NRAMP1 polymorphism with leprosy type but not susceptibility to leprosy per se in west Africans. Am. J. Trop. Med. Hyg. 65(2001)733-735.
12. Mira, M.T., Alcais, A., Di Pietrantonio, T., Thuc, N.V., Phuong, M.C., Abel, L. and Schurr, E. Segregation of HLA/TNF region is linked to leprosy clinical spectrum in families displaying mixed leprosy subtypes. Genes Immun. 4(2003)67-73.
13. Mira, M.T., Alcais, A., Van Thuc, N., Thai, V.H., Huong, N.T., Ba, N.N., Verner, A., Hudson, T.J., Abel, L. and Schurr, E. Chromosome 6q25 is linked to susceptibility to leprosy in a Vietnamese population. Nat Genet. 33(2003)412-415.
14. Roy, S., Frodsham, A., Saha, B., Hazra, S.K., Mascie-Taylor, C.G. and Hill, A.V. Association of vitamin D receptor genotype with leprosy type. J. Infect. Dis. 179(1999)187-191.
15. Roy, S., McGuire, W., Mascie-Taylor, C.G., Saha, B., Hazra, S.K., Hill, A.V. and Kwiatkowski, D. Tumor necrosis factor promoter polymorphism and susceptibility to lepromatous leprosy. J. Infect. Dis. 176(1997)530-532.
16. Shaw, M.A., Donaldson, I.J., Collins, A., Peacock, C.S., Lins-Lainson, Z., Shaw, J.J., Ramos, F., Silveira, F. and Blackwell, J.M. Association and linkage of leprosy phenotypes with HLA class II and tumour necrosis factor genes. Genes Immun. 2(2001)196-204.
17. Shields, E.D., Russell, D.A. and Pericak-Vance, M.A. Genetic epidemiology of the susceptibility to leprosy. J. Clin. Invest. 79(1987)1139-1143.
18. Siddiqui, M.R., Meisner, S., Tosh, K., Balkrishnan, K., Ghei, S., Fisher, S.E., Golding, M., Shanker Narayan, N.P., Sitaraman, T., Sengupta, U., Pitchappan, R. and Hill, A.V. A major susceptibility locus for leprosy in India maps to chromosome 10p13. Nat. Genet. 27(2001)439-441.
19. World Health Organization. Leprosy. Global situation. Wkly. Epidemiol. Rec. 77(2002)1-8.
1This is a Commentary on a recently published article by Mira, et al., the abstract of which can be found in the Current Literature section of this Journal on page 183.
Mailing address: Emil Skamene, Montreal General Hospital, Room A6-149, 1650 Cedar Avenue, Montreal, Que, H3G 1A4, Canada; Tele: (514) 937-6011 ext. 42434; FAX: (514) 933-7146; E-mail: emil.skamene@muhc.mcgill.ca