- Volume 65 , Number 4
- Page: 456–60
Testing candidate genes that may affect susceptibility to leprosy
ABSTRACT
Several statistical methods have been used to search familial data sets for marker alleles associated with the occurrence of a disease. In the present paper, a recently developed method is used to re-analyze published data on leprosy and candidate genes at the HLA loci. This new method of analysis, the randomization transmission disequilibrium test (TDT), confirmed previous conclusions that there was no significant evidence against random transmission at the HLA-A locus but significant positive association with the HLA-DR2 allele. The randomization TDT detected significant protective associations, that had not previously been found, with alleles HLA-B8 in Egyptian families and HLA-B21 (current nomenclature B*4901, 5001-5002) in South Indian families, highlighting a major advantage of permutation tests in analyzing candidate gene loci with rare alleles. These findings provide evidence that HLA class I restricted T lymphocytes may be of protective importance in leprosy.RÉSUMÉ
Différentes méthodes statistiques ont été utilisées pour rechercher, dans des séries de données familiales, les marqueurs alleles associés au développement d'une maladie. Dans le présent article, on utilise une méthode récemment développée pour réanalyser des données publiées sur la lèpre et les gènes candidats aux loci HLA. Cette nouvelle méthode d'analyse, le test de déséquilibre de la transmission aléatoire (TDT), a confirmé les conclusions précédentes, à savoir qu'il n'y avait aucune évidence significative contre la transmission aléatoire au locus HLA-A, mais bien une asociation positive significative avec I'allele HLA-DR2. Le TDT aléatoire a détecté des associations protectrices significatives qui n'avaient pas été découvertes auparavant, avec les alleles HLA-B8 dans des familles égyptiennes et HLA-B21 (nomenclature actuelle B*4901, 5001-5002) dans des families du Sud de l'Inde, mettant en évidence un avantage majeur des tests de permutation pour l'analyse des loci candidats avec des alleles rares. Ces découvertes montrent que les lymphocytes T HLA de classe I pourraient avoir une importance protectrice dans la lèpre.RESUMEN
Se han utilizado varios métodos estadísticos para buscar o identificar marcadores alélicos familiares asociados a diversas enfermedades. En el presente estudio se hizo uso de un método recientemente desarrollado para reanalizar los datos publicados sobre los genes HLA asociados con la lepra. Este nuevo método de análisis, la prueba de aleación de la transmisión del desequilibrio (TDT), confirmó la ausencia de asociación importante entre la enfermedad y el locus HLA-A y la asociación positiva entre la enfermedad y el alelo HLA-DR2. La prueba aleatoria TDT detectó nuevas asociaciones protectivas con el alelo HLA-B8 en familias egipcias y con el alelo HLA-B21 (nomenclatura actual B*4901, 5001-5002) en familias del sur de la India, señalando la ventaja de las pruebas de permutación en el análisis de los genes candidatos y su identifaciôn en alelos raros. Estos hallazgos proporcio-nan evidencias de que los linfocitos T restringidos por HLA clase I pueden ser de importancia protectora en la lepra.Leprosy is an infectious disease caused by a mycobacterium, Mycobacterium leprae. Individuals are known to respond differently to exposure to the bacterium, and this has suggested the possibility of some genetic contribution to individual susceptibility. The differential immune response to exposure is not only expressed by whether or not individuals develop leprosy, but also by differences in the subtype of leprosy that is expressed (19).
A genetic influence on the incidence of leprosy has been noted in twin studies (1,13,14). For a description of population or a case-control studies, see Todd, et al. (25) and Rani, et al . (16,17), and for more recent family-based studies, see de Vries," et al . (3); Fine, et al. (6); Wolf, et al. (28); van Eden, et al. (27); de Vries, et al. (4); Xu, et al . (29); Rawlinson, et al. (18) Levee, et al. (11) and Dessoukey, et al. (2). These studies revealed associations with different markers in the different populations. Nonrandom segregation of HLA markers has been observed for different types of leprosy in families from Surinam (3), China (29), India (4,6,29) and Egypt (2). There is no clear evidence about the actual mode of inheritance of the susceptibility and protective alleles, nor on how many loci are involved. HLA-DR2 is the marker allele that is most consistently implicated in the different studies.
In this paper, published familial data sets on leprosy will be re-analyzed using a recently developed statistical method highlighting the possibilities offered by this new test.
MATERIALS AND METHODS
The statistical method that will be used has the advantage over previous methods of allowing an analysis of multiple alleles at a locus that overcomes the problem of overestimation of statistical significance caused by multiple testing.
We shall assume a) that the genes at the HLA loci are candidate genes and not markers linked to candidate genes at nearby loci. This justifies an allelic study rather than a haplotypic segregation analysis or a method such as the LOD score, both of which search for linkage disequilibrium between a disease locus and a nearby marker locus; b) that no assumptions can be made about the number of genes involved, their origin or their levels of dominance and degrees of epistatic interaction. These assumptions lead to a choice between two methods of analysis.
The first method, developed by Sham and Curtis (21), models the probabilities that each allele will be transmitted from a parent to an affected child using logistic regression, but since the method relies on asymptotic results, it cannot safely be used to analyze data sets as small as those available for leprosy.
The second method of analysis, by Morris, et al. (15), is based on the transmission disequilibrium test (TDT) statistic (22), and can be used for any sample size. Schaid and Sommer (20) have presented a review of the statistical tests available to perform an analysis of familial data sets and have shown that the TDT statistic is the most robustly powerful statistic when the amount of dominance is unknown. The TDT statistic is a statistic calculated for each allele at a locus. For_allele A, say, all other alleles are labelled A. The total numberof times, b, that a heterozygous parent, A A, transmits the allele A to an affected offspring is compared with the numberof times, c, that the parent transmits an A allele using the McNemar chi-squared statistic: χ2 =(b - c)2/(b + c).
The Morris, et al. (15) analysis uses a randomization approach to test the significance of the largest χ2 value obtained from testing, in turn, each of the several alleles at the locus. The analysis avoids the loss of power associated with corrections for multiple testing and with the use of models that are over-elaborate and so involve too many unknown parameters. The TDT statistic is calculated for each of the marker alleles as described above and a significance level, p value, calculated for the largest of these TDT values. The randomization test is performed as follows: for each pair of parents in turn a random decision is made whether or not the transmitted and not transmitted alleles are exchanged. The largest TDT is then recorded for the new data set and the randomization is repeated a large number of times, for example, 2000 times. The p value is then calculated by counting how often the TDT values from the randomized data sets are larger than the observed value.
Homozygous parents are omitted since they cannot be used to indicate a tendency to transmit one allele rather than another to an affected child. The method can be extended to test sequentially for association with several alleles at a single locus (15) or with alleles at several loci.
The transmissions of linked marker alleles to different sibs within a sibship are not independent and so the usual tests cannot be applied to multiplex families. Fortunately, we are dealing with candidate genes not linked marker loci and, in this case, the tests are valid for multiplex families. All of the tests assume that the allele or alleles involved in the response to infection have the same effects in all the families.
Two published data sets will now be analyzed using the randomization method. In both publications, the selection criteria for the families were that at least two sibs were affected with leprosy in each family; there was at least one older unaffected sib and both parents were available for marker typing. The presence of an older unaffected sib is not required but should increase the power of the study by increasing the chance of heterozygosity in the parental genotypes.
The first data set (2) is based on 15 multiplex families from Cairo, Egypt. Information on the allelic forms present at the HLA-A, -B and -DR loci was available. At the HLA-A locus, 6 alleles were present: 1, 2, 3, 9, 11, 28. At the HLA-B locus, 13 alleles were present: 5, 7, 8, 12, 13, 14, 17, 18, 21, 27, 35, 37, 40. At the HLA-DR locus, 8 alleles were present: 1, 2, 3, 4, 5, 6, 7, 8.
The second data set was available from the literature but with arbitrary labelling of the alleles in each family (6). (Prof. Paul Fine kindly provided the fully identified data set.) The data are based on 72 multiplex families from South India. Information on the allelic forms present at HLA-A and -B loci were available. At the HLA-A locus, 13 markers were present: 1, 2, 3, 11, 23, 24, 26, 28, 29, 30, 31, 32, 33. At the HLA-B locus, 15 markers were present: 5, 7, 8, 12, 13, 15, 16, 17, 18, 21, 22, 27, 35, 37, 40. The current nomenclature for the A and B alleles where different from that used in the works referenced is as follows: A23 = A*2301; A28 = A*6801-6804, 6901; B5 = B*5101-5109, 5201; B12 = B*44024410, 4501; B15 = B*62, 63, 75, 76, 77; B16 = B*3801-3802, 3901-3910; B17 = B*5701-5704, 5801-5802; B21 = B*4901, 5001-5002; B22 = B*5401, 5501-5504, 5601-5603.
RESULTS
The data sets from Dessoukey and Fine have been analyzed using the method developed by Morris, et al. (15). The analysis was performed on all affected sibs with at least one heterozygous parent, this will be referred to as the AA group, as well as on sibs affected with a tuberculoid subtype of leprosy with at least one healthy heterozygous parent, this group will be referred to as the TH group. Some of the earlier studies indicated that statistical significance for particular alleles was only reached in one of these groups, justifying the analysis of these two groups separately.
At the A locus, there was no evidence against random transmission of any of the alleles.
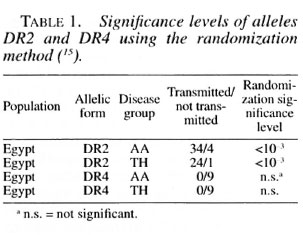
At the DR locus there was strong evidence for the nonrandom transmission of the candidate alleles. The association of disease with the HLA-DR2 allele was highly significant, confirming previously published results. The results are given in Table I for HLA-DR2 and also for HLA DR4. HLA-DR4 does not reach significance at the 5% level, possibly because HLA-DR4 is often found on the same haplotype as HLA-DR2 and the sequential randomization method first takes into account the presence of HLA-DR2.
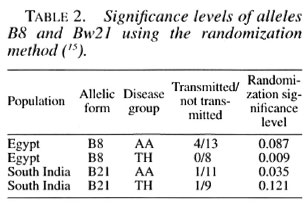
At the B locus, there was evidence against random transmission of the alleles (Table 2). A protective association with B8 was detected in the Egyptian families: but only reached significance in the TH group. A protective association with B21 was detected in the South Indian families that was statistically significant in the AA group. The fact that the two alleles B21 and B8 were relatively rare among the families could explain why they have not been mentioned in the earlier analysis of the same data and indicates a major advantage of the randomization method of analysis. As can be seen from Table 2, for each population the association with a particular allele is stronger in one disease group than in the other. But the actual degree of association varies little and at the 15% level every group is significant.
DISCUSSION
It is known that the HLA antigens encode for proteins which are involved in the immune response. Numerous associations and linkage studies have been reported for HLA types and infectious diseases. Leprosy has been studied by many investigators, for example, de Vries, et al. (3); van Eden, et al. (27) and reviewed by Todd, et al. (25). More recently, with an increasing understanding of the structural basis of HLA molecule functions (24) it has become possible to investigate the possible mechanisms of HLA association with infectious diseases. For example, the finding of an HLA-B53 association with resistance to severe malaria has provided evidence that HLA class I restricted T lymphocytes may be of importance in immune protection against this parasitic disease (9,10).
It is particularly interesting that here we have identified HLA class I associations with resistance to leprosy in addition to the well-known association with the HLA class II genes. Although HLA class I allelic associations have been suggested by some other studies in the past, reviewed by Todd, et al. (25), these were possible susceptibility associations. The evidence of protective associations provided here suggested that HLA class I restricted T lymphocytes may have a protective role against leprosy. Based on the increased susceptibility of mice lacking functional MHC class I molecules to M. tuberculosis, there has recently been substantial interest in a possible protective role for CD8+ class I restricted T lymphocytes in protection against tuberculosis (7). Recently, in mouse models of leprosy evidence for a possible protective role of CD8+ MHC class I restricted T cells has been found (12,23), and these results are consistent with such a role in human leprosy also.
Here we have identified different HLA class I alleles associated with protection against leprosy in different populations. In studies of other infectious diseases different HLA associations have been found in different populations, and it has been speculated that this may relate to genetic variation in infectious pathogens (8). The different types of leprosy probably involve immune responses that are qualitatively as well as quantitatively different, and so finding a particular HLA type associated only with one of the two polar types of leprosy is not surprising.
The particular advantage of the randomization test is that it allows a test for association with multiple alleles and can be used to analyze small data sets and to detect association with rare alleles. Unlike other methods, it does not rely on large sample distributional results. Clearly, further studies in the same and different populations are needed to confirm the conclusions from these small studies.
Acknowledgment. We thank Professor Paul Fine, London School of Hygiene and Tropical Medicine, for providing the data and for his very helpful comments and Professor Adrian Hill and Mr. Andrew Morris and the referees for helpful suggestions for improving an earlier version of this paper.
REFERENCES
1. CHAKRAVARTTI, M. R. and VoGEL, F. A twin study on leprosy. In: Topics in Human Genetics. Becker, P. E., Lenz, W., Vogel, F. and Wendt, G. G., eds. Stuttgart: Georg Thieme Publishers, 1973, pp. 1-30.
2. DESSOUKEY, M. W., EL-SHIENY, S. and SALLAM, T. HLA and leprosy: segregation and linkage study. Int. J. Dermatol. 35 (1996) 257-264.
3. DE VKIES, R. R. P., NIJENHUIS, L. E., LAI-A-FAT, R. F. M. and VAN ROOD, J. J. HLA-linked genetic control of host response to Mycobacterium leprae. Lancet 2 (1976) 1328-1330.
4. DE VRIES, R. R. P., MEIIRA. N. K., VAIDYA, M. C, GUPTE, M. D., KUAN, P. M. and VAN ROOD, J. J. HLA-linked control of susceptibility to tuberculoid leprosy and association of HLA-DR types. Tissue Antigens 16 (1980) 294-304.
5. DE VRIES, R. R. P., VAN EDEN, W. and OITHNHOFK, T. II. M. HLA Class II immune response genes and products in leprosy. Prog. Allergy 36 (1985) 95-113.
6. FINI:, P. E. M., WOLF, E., PRITCHARD, J., WATSON, B., BRADLEY, D. J., FESTENSTEIN, H. and CHACKO, C. J. G. HLA-Linked genes and leprosy: a family study in Karigiri, South India. J. Infect. Dis. 140 (1979) 152-161.
7. FI.YNN, J. L., GOLDSTEIN, M. M, TRIEHOLD, K. J., KOLI.LR, B. and BLOOM, B. R. Major histocompatibility complex class [-restricted T cells are required for resistance to Mycobacterium tuberculosis infection. Proc. Natl. Acad. Sci. U.S.A. 89(1992)12013-12017.
8. Hill., A. V. S. Genetic susceptibility to malaria and other infectious diseases: from the MHC to the whole genome. Parasitology 112 (1996) 1-10.
9. HILL, A. V., ALLSOPP, C. E., KWIATKOWSKI, D., ANSTEY, N. M., TWUMASI, P., ROWE, P. A., BENNETT, S., BREWSTER, D., MOMKTIAEL, A. J. and GREENWOOD, B. M. Common West African HLA antigens are associated with protection from severe malaria. Nature 352 (1991) 595-600.
10. HILL, A. V. S., ELVIN, J., WILLIS, A. C, AlDOO, M., ALLSOPP, C. E. M., GOTCH, F. M., GAO, X. M., TAKIGUCHI, M., GREENWOOD, B. M. and TOWN-SEND, A. R. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature 360 (1992) 434-439.
11. LEVEE, G., LIU, J., GICQUEL, B., CHANTEAU, S. and SCHURR, E. Genetic control of susceptibility to leprosy in French Polynesia; no evidence for linkage with markers on telomeric human chromosome 2. Int. J. Lepr. 62 (1994) 499-509.
12. MAW, W. W.; TOMIOKA, H., SATO, K., YAMADA, Y. and SAITO, H. Study on the roles of CD4+ and CD8+ T cells in the expression of host resistance to Mycobacterium leprae infection induced in athymic nude mice. Int. J. Lepr. 63 (1995) 539-545.
13. MOIIAMED AI.I, P- and RAMANUJAM, K. Genetics and leprosy: a study of leprosy in twins. Lepr. India 36 (1964) 77-86.
14. MOIIAMED AI.I, P. and RAMANUJAM, K. Leprosy in twins. Int. J. Lepr. 34 (1966) 405-407.
15. MORRIS, A. P., CURNOW, R. N. and WHITTAKER, J. C. Randomisation tests of disease marker associations. Ann. Hum. Genet. 61 (1997) 49-60.
16. RANI, R., FERNANDEZ-VINA, M. A., ZAHEER, S. A., BEKNA, K. R. and STASTNY, P. Study of HLA Class II alleles by PCR digotyping in leprosy patients from North India. Tissue Antigens 42 (1993) 1333-1337.
17. RANI, R., ZAHEER, S. A. and MUKHERJEE, R. DO human leukocyte antigens have a role to play in differential manifestation of multibacillary leprosy: a study on multibacillary leprosy patients from North India. Tissue Antigens 40 (1992) 124-127.
18. RAWLINSON, W. D., BASTEN, A., BRII ION, W. J. and SERJEANTSON, S. W. Leprosy and immunity: genetics and immune function in multiple case families. Immunol. Cell Biol. 66 (1988) 9-21.
19. RIDLEY, D. S. and JOPI.ING, W. H. Classification of leprosy according to immunity; a live-group system. Int. J. Lepr. 34 (1966) 255-273.
20. SCHAID, D. J. and SoMMER, S. S. Comparison of statistics for candidate-gene association studies using cases and parents. Am. J. Hum. Genet. 55 (1994)402-409.
21. SHAM, P. C. and CURTIS, D. An extended transmission/disequilibrium test (TDT) for multi-allele marker loci. Ann. Hum. Genet. 59 (1995) 323-336.
22. SPIELMAN, R. S., MGGINNIS, R. E. and EWENS, W.J. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mcllitus. Am. J. Hum. Genet. 52 (1993) 506-516.
23. STEINHOFF, U. and KAUIMANN, S. H. Specific lysis by CD8+ T cells of Schwann cells expressing Mycobacterium leprae antigens. Eur. J. Immunol. 18(1988)969-972.
24. STERN, L. I. and WILEY, D. C. Antigenic peptide binding by class I and class II histocompatibility proteins. Structure 2 (1994) 245-251.
25. TODD, J., WEST, B. C. and MCDONALD, J. C. HLA and leprosy: study in northern Louisiana and review. Rev. Infect. Dis. 12 (1990) 63-73.
26. VAN EDEN, W. and DE VRILS, R. R. P. HLA and leprosy: a re-evaluation. Lepr. Rev. 55 (1984) 89-104.
27. VAN EDEN, W., DE VRILS, R. R. P., MLHKA, N. K., VAIDYA, M. C, D'AMARO, J. and VAN ROOD, J. J. HLA segregation of tuberculoid leprosy: confirmation of the DR2 marker. J. Infect. Dis. 141 (1980) 693-701.
28. WOLF, E., FINE, P. E. M., PRITCHARD, J., WATSON, B., BRADLEY, D. J., FESTENSTEIN, H., CHACKO, C. J. G. and STEVENS, A. HLA-A, B and C antigens in South Indian families with leprosy. Tissue Antigens 15(1980) 436-446.
29. Xu, K., DE VRILS, R. R. P., FEI, H., VAN LLEUWEN, A., CHEN, R. and YE, G. HLA-linked control of predisposition to lepromatous leprosy. Int. J. Lepr. 53(1985)56-62.
1. M.Sc; Department of Applied Statistics, Harry Pitt Building, University of Reading, Whitekniehts Road, P.O. Box 240, Reading RG6 6FN. U.K.
2. Ph.D., Department of Applied Statistics, Harry Pitt Building, University of Reading, Whitekniehts Road, P.O. Box 240, Reading RG6 6FN. U.K.
Received for publication on 3 February 1997.
Accepted for publication in revised form on 11 August 1997.
Reprint requests to Prof. Curnow, 17 Pitts Lane, Farley, Reading, Berkshire RG6 1BX, U.K. or email: r.nc.urnow@reading.ac.uk