- Volume 64 , Number 4
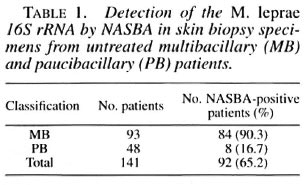
- Page: 396–403
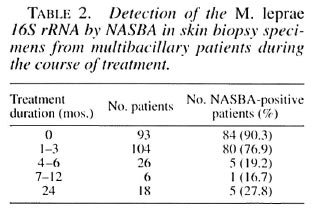
Use of NASBA® RNA amplification for detection of Mycobacterium leprae in skin biopsies f rom untreated and treated leprosy patients
ABSTRACT
This study was performed to assess the value of NASBA® RNA amplification of a 16S rRNA target for the detection of presumably viable Mycobacterium leprae in sections of skin biopsies f rom leprosy patients. The NASBA positivity rate was 90.4% (84/93) for untreated multibacillary (MB) patients [bacterial index (BI) > 2] and 16.7% (8/48) for the untreated paucibacillary (PB) patients (BI < 2). NASBA positivity showed a good concordance with the presence of solidly stained M. leprae [morphological index (MI)] in skin biopsies f rom leprosy patients, but no relationship could be demonstrated between the strength of the NASBA signals and the BI. Furthermore, the usefulness of the detection of 16S rRNA by NASBA to monitor the efficacy of leprosy treatment was investigated using an additional 154 biopsy specimens analyzed f rom 80 MB patients during the course of treatment. The NASBA positivity rate declined during treatment. A significant decrease was observed afteronly 1-3 months. Thèse results favor the view that détection of RNA by NASBA may reflect the viabil- ity of M. leprae.RÉSUMÉ
Cette étude a été réalisée pour estimer la valeur de l'amplification d'ARN par NASBA' d'une cible d'ARNr île 16S pour la détection de Mycobacterium leprae supposé viable dans des sections de biopsies cutanées provenant de malades de la lèpre. Le taux de positivité par NASBA était de 90.4% (84/93) pour des patients multibacillaires (MB) non traités [indice bactériologique (IB) > 2] et de 16.7% (8/48) pour des paucibacillaires (PB) non traités (IB < 2). La positivité du NASBA a montré une bonne corrélation avec la présence de M. leprae uniformément coloré [indice morphologique (IM)] dans les biopsies cutanées de malades de la lèpre, mais aucune relation n'a pu être mise en évidence entre la puissance des signaux du NASBA et I'IB. De plus, I'utilité de la détection de I'ARNs de 16S par NASBA pour suivre l'efficacité du traitement de la lèpre a été étudiée en analysant 154 échantillons supplémentaires de biopsies provenant de 80 malades MB durant leur traitement. Le taux de positivité du NASBA a diminué durant le traitement. Une diminution significative a été observée après seulement 1 à 3 mois. Ces résultats sont en faveur de l'idée que la détection d'ARN par le NASBA peut refléter la viabilité de M. leprae.RESUMEN
Este estudio se hizo para valorar la técnica de amplificación de rRNA 16s (RNA NASBA®) para establecer la viabilidad de Mycobacterium leprae en los coites de biopsias de piel de pacientes con lepra. La positividad de NASBA fue del 90.4% (84/93) para pacientes multibacilares (MB) no tratados (índice bacteriano. BI, de 2 o mayor) y del 16.79! (8/84) para pacientes paucibacilares no tratados (BI menor de 2). La positividad de NASBA mostró una buena concordancia con la presencia de bacilos sólidos en las biopsias de piel de los pacientes con lepra, pero no hubo relación entre la intensidad de las señales NASBA y el BI. También se investigó la utilidad de la técnica de detección de rRNA 16s por NASBA para evaluar la eficiencia del tratamiento antileproso usando 154 biopsias tomadas durante el curso de tratamiento de 80 pacientes con lepra MB. La positividad de NASBA declinó durante el tratamiento aunque una disminución significativa sólo se observó después de I a 3 meses de tratamiento. Los resultados apoyan la proposición de que la detección de RNA por NASBA pudiera reflejar la viabilidad de los bacilos en las lesiones de la lepra.Leprosy, caused by Mycobacterium leprae, is responsible for severe deformities, social stigma and economic loss. With the introduction of the short course World Health Organization multidrug therapy (WHO/MDT) in the early 1980s, the prevalence of the disease has been reduced dramatically. The number of leprosy patients in the world declined from an estimated 5.5 million in 1991 to 2.4 million in' 1994 (WHO press release, 1994). Recently, the WHO and its collaborators in the leprosy field have set for their goal the elimination of leprosy in the year 2000, elimination being defined by a worldwide prevalence of less than 1/10,000 (26). Despite the encouraging results with MDT, the incidence of leprosy remains at a stable level of 600,000 annually (WHO press release, 1994). Thus, early detection of new leprosy cases followed by adequate treatment will remain the core of leprosy control for many years to come.
A rapid method for quantifying viable M. leprae, an organism which still remains non-cultivatable in vitro, would be useful for assessing the efficacy and duration of existing and newly developed chemotherapeutic regimens. In addition, such a method for detection of viable mycobacteria would have the potential for differential diagnosis of a relapse and a reversal reaction. Furthermore, it could be utilized to assess subclinical infection with M. leprae. The mouse foot pad assay (MFA) is a well established and useful method for measuring the viability of M. leprae in vivo (4, 16)- However, MFA is insensitive, time consuming and very laborious. In the past, several rapid but indirect techniques have been proposed for the determination of the viability of M. leprae in vitro, such as the morphological index (MI) determination (21), fluorescent stainings (13), the measurement of the ATP content (7) and, more recently, laser micro-probe mass spectrometry (LAMMS) (15). However, all of these assays have limitations in terms of either sensitivity or objectivity or they are not easy to perform (2, 10).
Based on the notion that nucleic acids (NA) are present in viable mycobacteria and are vulnerable to degradation upon bacterial death, the polymerase chain reaction (PCR) for the detection of DNA has been employed as a method for monitoring the efficacy of treatment (6,9,23,24). We have employed nucleic acid sequence-based amplification (NASBA) (5) for the detection of 16S rRNA as a method to assess mycobacterial viability based on the presumption that 16S rRNA is directly associated with bacterial metabolic activity (11,18). In this study, we present a standardized protocol for NASBA analysis for the detection and identification of M. leprae 16S rRNA in skin biopsies. We used this protocol to monitor the efficiency of treatment in patients with leprosy and compared NASBA results with bacteriological findings.
MATERIALS AND METHODS
Skin biopsies. A total of 301 biopsy specimens from skin lesions were collected from leprosy patients residing in The Philippines, Thailand and The Netherlands. Patients had been classified as multibacillary (MB) when the mean bacterial index (BI), based on the enumeration of acid-fast bacteria (AFB) in slit-skin smears, was > 2 and classified as paucibacillary (PB) when the Bl was < 2. Ninety-five skin biopsies were collected from untreated MB patients and 48 from untreated PB patients. In addition, 158 biopsy specimens were collected from 80 MB patients at different times after the start of MDT. As controls, skin biopsies from seven patients with skin diseases other than leprosy, such as sarcoidosis and leishmaniasis, were included as well as specimens from four patients with normal skin undergoing reconstructive surgery. Parts of the 4-mm punch biopsy specimens were frozen and sectioned as described by Yoon, et al. (27), the others were homogenized in 5 ml of Hanks' balanced salt solution (HBSS). The frozen biopsy specimens were stored at -70ºC and transported on dry ice; these specimens were further processed as described below. From the biopsy homogenates in HBSS, 50 µ l was directly added to 950 µ l of L6-buffer (1) and thereafter stored at -70ºC. No significant differences between the NASBA results from frozen and homogenated biopsy specimens were found (all comparisons by chi-squared test: p > 0.05) and, therefore, the NASBA results of all the specimens were analyzed together.
BI and morphology. The BI was determined by enumeration of AFB on sections in the granuloma after staining by the modified Fite method (14). The morphological index (Ml) of the M. leprae was determined according to Waters and Rees and stained bacteria were classified either as solid (uniform staining), fragmented (irregularly stained but intact bacteria), or granular (no intact bacteria). A positive MI was defined by the presence of solidly stained bacteria (> 0%).
Specimen preparation. From each frozen biopsy specimen, 5 sections of 5 pm were cut and put into a conical screw-cap vial as described by Yoon, et al. (24). Upon arrival of the frozen sections of the skin biopsies at the Royal Tropical Institute in Amsterdam, 1 ml of L6- buffer (1) was directly added to the tubes and approximately 100-200 µ l of 0.1-mm diameter zirconium beads (Biospec Products, Bartlesville, Oklahoma, U.S.A.) were put into each tube. The tubes were shaken in a Minibeadbeater (MiniBeadBeater model 3110; Biospec) for 220 sec. The lysis of the mycobacteria in these specimens and the biopsy homogenated in L6-buffer was facilitated by freeze/boiling (-70oC/100ºC) two times with an interval of 5 min. After the last boiling step, the suspensions containing beads were shaken again in the Minibead beater for 120 sec. Afterward these tubes were cenfrifuged (13,000 rpm) for 5 min to settle the beads and, subsequently, the supernatants were transferred to new tubes and stored at -70ºC until NA extraction was performed.
NA isolation. The NA were purified by the guanidinium thiocyanate method using silica as described by Boom, et al. (1). The NA isolate was stored at -20ºC until further use. Two µ l of the NA isolate was used for NASBA.
Amplification, hybridization and detection. NASBA was performed as described previously (18) with minor modifications. The final volume of the reaction mixture was 25 µ l to which a 5- µ l template was added. The final concentration of the reaction mixture was 40 mM Tris/HCl, pH 8.5; 12 mM MgCl2 70 mM KC1; 5 mM DTT; 1 mM of each dNTP; 2 mM of ATP, CTP, UTP; 1.5 mM of GTP and 0.5 mM ITP, 15% (v/v) DMSO, 0.2 mM OT727 and OT737 (19). An enzyme mixture was assembled so that the final concentration of the NASBA reaction was composed of 0.1 µ g/ µ l BSA, 125 mM Sorbitol (Pharmacia, Uppsala, Sweden), 5 mM DTT, 40 U T7 RNA polymerase (Pharmacia), 8 U AMVRT (Seikagaku America, St. Petersburg, Florida, U.S.A.) and 0.1 U RNAse H (Pharmacia). Isothermal amplification of the RNA target was performed by incubation of these samples at 41 ºC for 1.5 hr.
Detection of the amplified RNA was done according to the electrochemiluminescence (ECL) technique with minor modifications: 5 µ l of NASBA product was diluted 100-fold in RNase-free water. The 5'-biotinyIated probe (5'-bio-OT1490 18) was used to capture the NASBA product. For the identification of the NASBA products, the M. leprae detection probe (5'TAGGA CTTCA AGGCG CATGT-3'; OT1681) was used (18). The label used for ECL-detection was a tris [2,2-bipyridine] ruthenium [II] complex. This label emits light as a result of chemical reactions taking place at the surface of an electrode. This signal can be quantified with a dynamic range over 5 orders of magnitude using a specifically developed detection instrument. To increase the specificity of this probe, it was mixed 1:100 with the four unlabeled probes containing the species-specific 16S rRNA sequence for M. smegmatis, M. tuberculosis, M. avium/paratuberculosis, and M. intracellulare (18). The cutoff value was calculated as the mean value of the negative controls (N = 10) plus three standard deviations, being 3000. The entire procedure from isolation to amplification and detection can be performed in 1 day.
Controls for amplification. One negative control for every five clinical specimens was included in order to assess carryover contamination during NA isolation and amplification. These control samples containing only water were randomly distributed in between and processed along with the clinical specimens. The presence of inhibiting substances in the NA isolate affecting the amplification reaction was monitored by spiking those samples with a negative NASBA signal with 103 molecules of in vitro synthesized RNA containing the sequence of 16S rRNA of M. smegmatis (18) .
Analysis of NASBA. When a positive NASBA signal was obtained, the presence of M. leprae in the skin biopsy was considered. A sample was considered negative when it did not show amplification in NASBA and did not inhibit the amplification of the spiked amplification reaction. Skin biopsies containing inhibiting substances (N = 6) for the amplification reaction were excluded from our analysis.
RESULTS
The detection of M. leprae 16S rRNA by NASBA in skin biopsies was assessed by analyzing specimens collected from 141 untreated leprosy patients: 93 MB and 48 PB patients (Table 1). The positivity rate of NASBA for the MB specimens was 90.3% and for the PB patients, 16.7%. The 11 skin biopsies from 7 patients with skin diseases other than leprosy and 4 patients undergoing reconstructive surgery were till negative (results not shown).
The Figure shows the numerical data of NASBA as found in the FCL detection assay in relation to the histopathological BI of 76 skin biopsies from 52 MB and 24 PB untreated patients. The majority of specimens with a BI of < 2 were negative in NASBA, except one, while specimens with a BI of > 2 were mostly positive, except five specimens. A wide range of NASBA signals was observed, but no relation between the strength of the NASBA signals and the histopathological BI could be demonstrated.
The usefulness of the detection of 16S rRNA by NASBA to monitor the efficacy of leprosy treatment was investigated using an additional 154 biopsy specimens collected from 80 MB patients during the course of treatment. As shown in Table 2, the NASBA positivity rate declined during treatment. A significant decrease was observed after only 1-3 months (compared to the specimens from untreated patients; p = 0.01, chi-squared test) and again after 4-6 months of treatment (compared to the specimens from patients treated for 1-3 months; p < 0.0001, chi-squared test). Five of the 18 MB patients (27.8%) showed a positive NASBA signal after 24 months of MDT. From 2 of these 5 patients further clinical data were available. One of these patients had a history of noncompliance with therapy; the other was suspected for a relapse of his leprosy based on clinical data. Furthermore, this latter patient showed consistent high levels of anti-phenolic glycolipid-I (PGL-I) antibody during the entire course of treatment (data not shown).
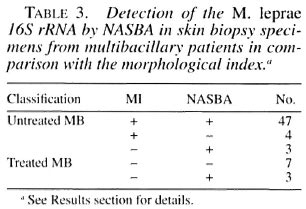
To further investigate whether the detection of 16S rRNA is an indicator of potentially viable bacteria, the relationship between NASBA and the morphology of the stained mycobacteria was investigated (Table 3). From 54 untreated MB patients, the MI was available for analysis. Fifty-one of these patients had a positive MI of which 47 (87%) were positive in NASBA. The correlation between the strength of the ECL signals and the MI was r = 0.22. Four specimens with a positive MI had a negative NASBA; these patients had a BI varying from 3 to 5 and were also negative in PCR based on the detection of a DNA sequence of the pra gene (results not shown21) . The biopsy specimens of the three patients with an MI of 0 were positive in NASBA; these specimens, till with a histopathological BI of 5, were also positive in a PCR. In addition, from 10 MB patients on treatment (duration from 3 months to 24 months) with an MI of 0, NASBA was positive in three. Two of those three patients were on treatment for only 3 months and both had a histopathological BI of 4. One MI-negative NASBA-positive specimen was from a patient who was on treatment for 12 months, and who w;ts suspected of having a relapse based on clinical findings.
DISCUSSION
Since M. leprae is non-cultivatable in vitro, viability assessment and thus the monitoring of chemotherapy has to rely on indirect methods which are not sensitive and/or easy to perform. Since ribosomal disappearance is one of the first ultrastructural signs associated with the loss of mycobacterial viability (17), the presence of I6S rRNA may indicate active metabolism and/or replication. Based on this assumption, we have employed previously the rapid and relatively simple RNA amplification method NASBA for the accurate detection and identification of low numbers of mycobacteria in order to assess the viability (19). We have previously shown that RNA detection by NASBA indicates the presence of viable mycobacteria in vitro (18). In this study, we used NASBA for the detection of M. leprae in skin biopsies from untreated and treated leprosy patients, and speculate that it has the potential to assess the viability of M. leprae.
The results of NASBA applied to skin biopsies from untreated MB patients showed a high positive rate (90.3%), but biopsies from untreated PB patients had a low positive NASBA rate (16.7%). This sensitivity, high for MB patients and low for PB patients, is comparable with existing diagnostic techniques, including PCR and serology (6,12,24). Only 1% of the total bacterial load, being 107 to 108 (dead and viable), in each PB patient is viable (25). Therefore, there is only a small chance to detect viable M. leprae in a few sections from a skin biopsy and, thus, failure to detect the presence of M. leprae in PB patients by NASBA could be the consequence of the small fraction of the specimen subjected to testing.
The strength of the NASBA/ECL signals did not correlate with either the histopathological BI or the MI values. This may be due to the intrinsic properties of this RNA amplification technique. Amplification efficiency in NASBA varies inversely with the numbers of input RNA molecules used, with greater efficiency resulting from a small number of RNA templates (5). As a consequence, NASBA behaves more like an on/off reaction (The Figure) and no quantitative extrapolations from the NASBA/ ECL signals can be made to the number of RNA molecules in the specimen, unless the test is specifically adjusted for that purpose (20). Nevertheless, the qualitative NASBA results showed good concordance with the MI (Table 3). Only 4 of the 51 specimens (7.8%) with a positive MI were negative in NASBA. However, since the same specimens of these untreated MB patients were also negative in a PCR assay, it is unlikely that this discrepancy was caused by a technical error in the NASBA procedure. More likely, the MI may overestimate the number of viable bacteria, due to the time lag between cell death and degenerative changes that result in irregular staining (9). Moreover, the MI determination and the NASBA were not performed on the same section of the skin biopsy.
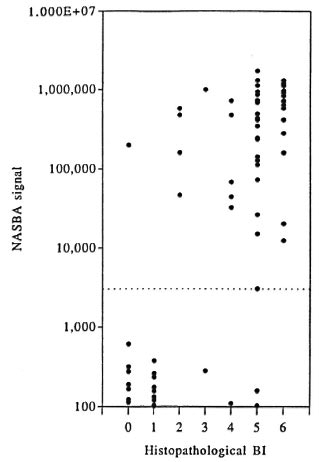
The Figure: Strength of NASBAa signals in relation to the histophathological BI of 76 sections of skin biopsy speciments from 76 untreated leprosy patients.
In three specimens from untreated and three from treated patients, no solid-stained M. leprae could be identified, while the NASBA on these specimens was positive (Table 3). It is unlikely that this is the result of contamination of the NASBA, since special care was taken during specimen processing; negative controls were processed along with all patient specimens and these remained negative throughout the study. It is more likely that in these cases the number of solid-stained M. leprae was underrated. Although the MI is a good indication of the viability of the mycobacteria (22), it should be noted that in case of low numbers of mycobacteria present in the section, the quantitative outcome of the MI is neither a very reliable nor a very sensitive measure of viability (9).
The idea that RNA degradation occurs rapidly upon cell death and that RNA detection by NASBA may, thus, indicate the presence of viable M. leprae, was illustrated by the rapid decline of NASBA positivity during the course of treatment of MB patients (Table 2). It is not likely that the decrease in the NASBA positivity is related to a decrease in the bacillary load at the site of the biopsy. In general, BI declines at a slower pace and, as discussed above, NASBA was more related to the MI than to the BI. Our findings are in agreement with current concepts of the effect of leprosy treatment: 99.999% of the viable M. leprae are killed by a few doses of rifampin (8). A low number of bacteria that remain viable, the so-called "persisters." is maintained during treatment (3). It has been suggested that such persisting mycobacteria are in a dormant state during the course of leprosy treatment and that their elimination depends upon the host's ability to clear these microorganisms (8). The potential of NASBA to measure leprosy treatment failures is illustrated by two patients with positive NASBA signals at the end of their treatment. One treatment failure could be attributed to the patient's lack of compliance and the other to a relapse of infection due to unknown reasons. The applicability of NASBA to assess the viability of M. leprae in conjunction with efficacy of treatment of individual leprosy patients will be explored in a prospective study.
In conclusion. NASBA is a rapid and relatively simple tool for the specific and sensitive defection of mycobacterial RNA. The results presented here support our premise that the detection of RNA indicates the presence of viable M. leprae in skin biopsies of leprosy patients. However, viability studies in mouse foot pads are required to further sustain our hypothesis. It would be of interest to investigate whether the presence of viable M. leprae at the time of diagnosis has any prognostic value for individual patients in relation to response to leprosy treatment. If, indeed, detection of mycobacterial RNA provides a rapid means of viability assessment, then NASBA provides a variety of clinical and research applications for the detection of viable M. leprae, for instance, in detecting persisting and drug-resistant mycobacteria and in tracking sources of transmission.
Acknowledgment. The authors wish to thank ail the staff members (R. V. Cellona, Ma. V. F. Balagon, L. G. Villahermosa, T. T. Fajardo, Jr., and R. M. Abalos) of the Leonard Wood Memorial Center tor clinical and histopathological assessments of the patients. Also, the help of C.E. Verhagen and A.A.M. Bulling in cutting the skin biopsies is greatly appreciated.
This investigation received financial support from the QM Gastmann-Wichers Foundation, the UNDP/ World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR), the Netherlands Leprosy Relief Association (NSL), and the Commission of the European Communities, Directorate General for Science, Research and Development (grant TS3-CT91-0036), and ALM International support provided to the Leonard Wood Memorial/American Leprosy Foundation.
REFERENCES
1. BOOM, R., SOL, C. J. A., SALIMANS, M. M. M., JANSEN, C. L., WEKTHEIM-VAN DlLLEN, P. M. E. and VAN DER NOORDAA, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28(1990)459-503.
2. CHANG, Y. T. Are all nonsolid Mycobacterium leprae dead? Does a negative finding in the mouse foot pad indicate that there is actually no growth of M. leprae in the animals? Int. J. Lepr. 45(1976)235-240.
3. COLSTON, M. J., ELLARD, G. A., FERRACCI, C. A. P., GROSSET, J. H., GROSSETETE, G., IYER, C. G. S., JACOBSON, R. R., Jr, B., LWIN, K., LEIKER, D. L., LEVY, L, NOORDEEN, S. K. and PATTYN, S. R. Persisting Mycobacterium leprae among THELEP trial patients in Bamako and Chigleput. Lepr. Rev. 58(1987)325-337.
4. COLSTON, M. J., HILSON, G. R. F. and BANERJEE, D. K. The "proportional bactericidal test": a method for assessing bactericidal activity of drugs against Mycobacterium leprae in mice. Lepr. Rev. 49(1978)7-15.
5. COMPTON, J. Nucleic acid sequence-based amplification. Nature 350(1991)91-92.
6. DE WIT, M. Y. L., FABER, W. R., KRIEG, S. R., DOUGLAS, J. T, LUCAS, S. B., MONTREEWASUWAT, N., PATTYN, S. R., HUSSAIN, R., PONNIGHAUS, J. M., HARTSKEERL, R. A. and KLATSER, P. R. Application of a polymerase chain reaction for the detection of Mycobacterium leprae in skin tissues. J. Clin. Microbiol. 29(1991)906-910.
7. DHOPLE, A. M. Adenosine triphosphate content of Mycobacterium leprae from leprosy patients. Int. J. Lepr. 52(1983)183-188.
8. GROSSET, J.-H. Progress in the chemotherapy of leprosy. Int. J. Lepr. 62(1994)268-277.
9. JAMIL, S., KEER, J. T, LUCAS, S. B., DOCKRELL, H. M., CHIANG, T. J., HUSSAIN, R. and STOKER, N. G. Use of polymerase chain reaction to assess efficacy of leprosy chemotherapy. Lancet 342(1993)264-268.
10. KATOCH, V. M., KATOCH, K., RAMANATHAN, U., SHARMA, V. D., SHIVANNAVAR, C. T, DATTA, A. K. and BHARADWAJ, V. P. Effect of chemotherapy on viability of Mycobacterium leprae as determined by ATP content, morphological index and FDAEB fluorescent staining. Int. J. Lepr. 57(1988)615-621.
11. KAWA, D. E., PENNELL, D. R., KUBISTA, L. N. and SCHELL, R. F. Development of a rapid method for determining the susceptibility of Mycobacterium tuberculosis to isoniazid using the gen-probe DNA hybridization system. Antimicrob. Agents Chemother. 33(1989)1000-1005.
12. KLATSER, P. R. Serology of leprosy. Trop. Geogr. Med. 46(1994)115-118.
13. KVACH, J. T. and J. R. VEKAS. A fluorescent staining procedure for determining the viability of mycobacterial cells. Int. J. Lepr. 50(1982)183-192.
14. RIDLEY, R. S. Pathogenesis of Leprosy ami Related Diseases, London: John Wright, 1988, p. 240.
15. SEYDEL, U., HAAS, M., RIETSCHEL, E. T. and LINDNER, B. Laser microprobe mass spectrometry of individual bacterial organisms and of isolated bacterial compounds: a tool in microbiology. J. Microbiol. Meth. 15(1992)167-183.
16. SHEPARD, C. C. A kinetic method for the study of activity of drugs against Mycobacterium leprae in mice. Int. J. Lepr. 35(1967)429-454.
17. SILVA, M. T, APPELBERG, R., SILVA, N. T. and M.ACEDO, P. M. In vivo killing and degradation of Mycobacterium aurum within mouse peritoneal macrophages. Infect. Immun. 55(1987)2006-2016.
18. VAN DER VLIET, G. M. E., SCHEPERS, P., SCHUKKINK, R. A. F., VAN GEMEN, B. and KLATSER, P. R. Assessment of mycobacterial viability through RNA-amplification. Antimicrob. Agents Chemother. 38(1994)1959-1965.
19. VAN DER VLIET, G. M. E., SCHUKKINK, R. A. F., VAN GEMEN, B., SCHEPER, P. and KLATSER, P. R. Nucleic acid sequence-based amplification (NASBA) for the identification of mycobacteria. J. Gen. Microbiol. 139(1993)2423-2429.
20. VAN GEMEN, B., VAN BEUNINGEN, R., NABBE, A., VAN STRIJP, D., JURRIAANS, S., LENS, P. and KIEVITS, T. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J. Virol. Methods 49(1994)157-168.
21. WATERS, M. F. R. and REES, R. J. W. Changes in the morphology of Mycobacterium leprae in patients under treatment. Int. J. Lepr. 30(1962)266-277.
22. WELCH, T. M., GELBER. R. H., MURRAY, L. P., O'NEILL, S. M. and LEVY, L. Viability of Mycobacterium leprae after multiplication in mice. Infect. Immun. 30(1980)325-328.
23. WlCHITWECHKARN, J., KARNJAN, S., SHUNTAWUTTISEITEE, S., SORNPRASIT, C, KAMPIRAPAP, K. and PEERAPAKORN, S. Detection of Mycobacterium leprae infection by PCR. J. Clin. Microbiol. 33(1995)45-49.
24. WILLIAMS, D. L., GILLIS, T. P., FIALLO, P., JOB, C. K., GELBER, R. H., HILL, C. and IZUMI, S. Detection of Mycobacterium leprae and the potential tor monitoring antileprosy drug therapy directly from skin biopsies by PCR. Mol. Cell Probes 6(1992)401-410.
25. WHO COMMITTEE ON EXPERIMENTAL CHEMOTHERAPY. Experimental chemotherapy in leprosy. Bull. WHO 53(1976)425-433.
26. WORLD HEALTH ORGANIZATION. Eliminating leprosy as a public health problem before the year 2000 . Geneva: World Health Organization, 1994.
27. YOON, K.-H., CHO, S.-N., LEE, M.-K., ABALOS, R. M., CELLONA, R. V., FAJARDO, T. T., JR., GUIDO, L. S., DELÀ CRUZ, E. C, WALSH, G. P. and KIM, J.-D. Evaluation of the polymerase chain reaction amplification of Mycobacterium leprae -specific repetitive sequence in biopsy specimens from leprosy patients. J. Clin. Microbiol. 31(1993)895-899.
1. Ph.D., Royal Tropical Institute, Department of Biomedical Research, Meibergdreef 39. 1105 AZ Amsterdam, The Netherlands.
2. Ph.D., Yonsei University, College of Medicine, Department of Microbiology, 134 Shinchondong Seodaemoonku, Seoul 120752, Korea.
3. M.D., Sasakawa Research Building, Soi Bamrasnaradoon Hospital, Tiwanond Road, Nonthaburi Province 11000, Thailand.
4. Ing., Royal Tropical Institute, Department of Biomedical Research, Meibergdreef 39. 1105 AZ Amsterdam, The Netherlands.
5. Ing., Organon Teknika, Boseind 15. 5281 RM Boxtel, The Netherlands.
6. Ph.D., Organon Teknika, Boseind 15. 5281 RM Boxtel, The Netherlands.
7. Ph.D., University of Amsterdam, Depts. of Dermatology and Pathology, Academic Medical Center, Meiberedreef 9, 1105 AZ Amsterdam, The Netherlands.
8. M.D., University of Amsterdam, Dept. of Dermatology, Academic Medical Center, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands.
9. Ph.D., Leonard Wood Memorial Leprosy Research Center, P.O. Box 727, Cebu City, The Philippines.
10. Ph.D., Royal Tropical Institute, Department of Biomedical Research, Meibergdreef 39. 1105 AZ Amsterdam, The Netherlands.
Reprint requests to Dr. Klatser at the above address or FAX 31-20-697-1841.
Received for publication on 25 March 1996.
Accepted for publication in revised form on 24 July 1996.