- Volume 64 , Number 4
- Page: 404–8
Polymerase chain reaction of nasal swabs f rom tuberculosis patients and their contacts
ABSTRACT
Previous studies have found Mycobacterium leprae in nasal swabs f rom leprosy patients, their contacts, and persons living in endemic areas. It might be expected that M. tuberculosis would be present on nasal mucosa of pulmonary tuberculosis patients, but whether they can be detected in patients or contacts is unknown. We used the polymerase chain reaction (PCR) technique on nasal swabs f rom tuberculosis patients, contacts of tuberculosis patients, leprosy patients, and London controls to look for both M. tuberculosis and M. leprae. Swabs dipped in sputum specimens f rom smear-positive patients were used as positive controls. The PCRs were conducted in two independent laboratories. M. tuberculosis was detected in nasal swabs f rom 6/16 smear-positive tuberculosis patients and f rom 1/10 household contacts by one of the laboratories. All of the sputum swabs were positive for M. tuberculosis, and all of the London controls were negative. M. leprae were found in nasal swabs f rom 2/5 leprosy patients, but one laboratory also reported M. leprae in swabs f rom 4/21 tuberculosis patients and f rom one sputum specimen. The results show that M. tuberculosis can be found in the noses of some tuberculosis patients, and suggest that the bacilli also may be detected in some household contacts.The comparisons with M. leprae and between the two laboratories give further in- sights into the sensitivity and specificity of the technique.RÉSUMÉ
Des éludes antérieures ont trouvé des Mycobacterium leprae dans les décharges nasales de malades de la lèpre, de leurs contacts, et de personnes vivant en régions endémiques. On pourrait s'attendre à ce que M. tuberculosis soit présent sur les muqueuses nasales de patients présentant une tuberculose pulmonaire, mais on ne sait pas s'il peut être détecté chez les patients ou des contacts. Nous avons utilisé la technique de la réaction de polymerase en chaîne (PCR) sur des produits de décharge nasale provenant de patients tuberculeux, des contacts de patients tuberculeux, des patients lépreux et des témoins de la ville de Londres pour rechercher M. tuberculosis et M. Leprae. Des cotons plongés dans îles spécimens de crachats provenant de patients dont les expectorations étaient positives à l'examen direct ont été utilisés comme contrôles positifs. Les PCR ont été réalisées dans deux laboratoires indépendants. M. tuberculosis a été détecté dans les décharges nasales de 6 patients tuberculeux sur 16 positifs à l'examen des expectorations, et chez un contact domiciliaire sur dix par l'un des laboratoires. Tous les cotons imbibés d'expectorations étaient positifs pour M. tuberculosis, et tous les contrôles londoniens étaient négatifs. M, leprae a été trouvé dans les décharges nasales de deux patients lépreux sur cinq, mais un laboratoire a aussi rapporté du M. leprae dans les décharges de 4 patients tuberculeux sur 21 ainsi que d'un échantillon d'expectorations. Les résultats montrent que M. tuberculosis peut être trouvé dans le nez, de certains patients tuberculeux, et suggèrent que les bacilles peuvent aussi être détectés chez certains contacts domiciliaires. Les comparaisons avec M. leprae et entre les deux laboratoires donnent des informations supplémentaires quant à la sensibilité et la spécificité de la technique.RESUMEN
En estudios previos ya se ha reportado la presencia de Mycobacterium leprae en los exudados nasales délos pacientes con lepra, en los de sus contactos, y en personas que viven en áreas endémicas. Podría esperarse que M. tuberculosis estuviera presente en la mucosa nasal de los pacientes con tuberculosis pulmonar o en sus contactos, pero esto hasta ahora no se había estudiado. Nosotros usamos la reacción en cadena de la polimerasa (PCR) para buscar tanto M. tuberculosis como M. leprae en los exudados nasales tic pacientes con tuberculosis, en sus contactos, en pacientes con lepra, y en controles londinenses. Como controles positivos se usaron hisopos sumergidos en el esputo de pacientes bacilíferos. Los PCRs se hicieron en 2 laboratorios independientes. Uno de los laboratorios detectó M. tuberculosis en los exudados de 6 de 16 pacientes con tuberculosis BAAR positivos y en 10 de sus contactos convivientes. Todas las muestras de esputo fueron positivas para M. tuberculosis y todos los controles londinenses fueron negativos. Dos de 5 pacientes con lepra tuvieron M. leprae en sus exudados pero un laboratorio también reportó M. leprae en los exudados de 4 de 21 pacientes con tuberculosis y en una muestra tic esputo. Los resultados muestran que M. tuberculosis puede encontrarse en la nariz de algunos pacientes con tuberculosis y en algunos de sus contactos convivientes. La comparación de los hallazgos con M. leprae y de los resultados de los 2 laboratorios, proporciona información adicinal sobre la sensibilidad y la especificidad de la técnica.Several studies using the polymerase chain reaction (PCR) technique have identified leprosy bacilli in nasal swabs from leprosy patients, their contacts, and other persons living in endemic areas (3,5,9). The nose is a site of carriage and shedding of Mycobacterium leprae, and a possible portal of entry. Nasal tuberculous lesions are rare in humans (9,10) but M. bovis has been isolated from the nasal secretions of infected cattle (2,8). Given the large numbers of bacilli coughed up from the lungs of patients with pulmonary tuberculosis, one might expect that some bacilli would be present in the mucosa of the upper respiratory tract, including the nasal cavity. The nose acts as an air filter, and so inhaled bacilli may concentrate there and be detectable, whether or not they multiply in the nasal cavity. Detection of M. tuberculosis in the noses of patients and contacts could give us clues to the transmission of tubercle bacilli in different environments.
In this pilot study in northern Malawi, nasal swabs were taken from tuberculosis patients, their contacts and various controls, and tested for both M. tuberculosis and M. leprae in laboratories in London and Amsterdam.
MATERIALS AND METHODS
Nasal swabs were taken from each nostril of newly diagnosed, pulmonary tuberculous inpatients at Karonga District Hospital, northern Malawi. The sterile, cotton wool swabs were dipped in sterile saline before use and afterward were stored at -20ºC. They were transported to Europe in cold boxes with ice. Nasal swabs were also taken from household contacts of three of the smear-positive patients, from healthy volunteers in London ("negative controls"), from the medical officer after a tuberculosis ward round in Malawi, and from leprosy patients. "Positive control" swabs were dipped in the sputum of smear-positive tuberculosis patients. Half of the swabs from each patient were sent to each of the two laboratories. The laboratories were blinded as to the source of the swabs.
Laboratory methods
Laboratory A. The target for the tuberculosis polymerase chain reaction (PCR) was the IS6110 insertion sequence; for M. leprae, it was the pra gene sequence. Treatment of the swab specimens with lysis buffer (5) and PCR (5,7) was carried out as described previously. Six tubes of lysis buffer were used as negative controls in each run. Positive amplification, as judged by agarose gel electrophoresis, was confirmed by hybridization. Negative samples were screened for inhibition of the PCR by spiking with a modified template for the M. leprae PCR (3) and with M. tuberculosis DNA for the M. tuberculosis PCR. When inhibition was found the sample was purified and retested as previously described (3,7) . Each extract was amplified twice to confirm the results, and the amplifications were repeated if they were not concordant.
Laboratory B. The target for the tuberculosis PCR was the IS6110 insertion sequence; for M. leprae, it was the RLEP sequence. DNA extraction and PCR were carried out as described previously (4) except that individual swabs were resuspended in 100 µl PCR buffer containing 100 µ g/ml proteinase K and 0.5% Tween 20, and incubated at 60ºC for 1 hr prior to freeze-boiling. For each batch of samples five control specimens were prepared and treated identically to the test specimens except for the addition of swabs. PCR was carried out using a hot-start protocol. Samples were screened for the presence of inhibitors using PCR with amplifiable concentrations of M. tuberculosis DNA and primers specific for M. tuberculosis DNA in the extracts. Ten further negative controls (reaction mixture) were used in each run. PCR products were detected using colorimetric methods. Each extract was amplified twice to confirm the results, and this was repeated if they were not concordant.
RESULTS
Eighty nasal swabs for each laboratory were taken from 44 subjects. Eight additional sets of swabs were dipped in sputum specimens from smear-positive tuberculosis patients. The number of swabs from each source and the results of the PCR studies are shown Table 1. Results were undetermined in 14 specimens in Laboratory B and for one in Laboratory A due to the presence of inhibitors.
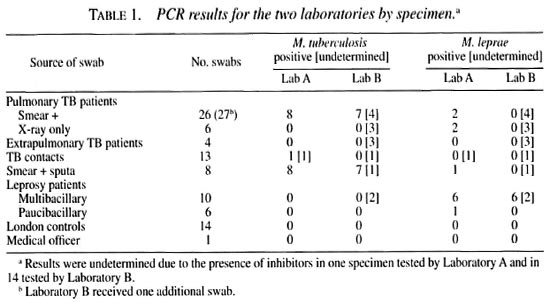
The results are shown by patient in Table 2. All patients contributed one or two nasal swabs for each laboratory, except for two of the leprosy cases. One multibacillary (MB) leprosy case had six specimens taken, all of which were positive for M. leprae and negative for M. tuberculosis in both laboratories; one paucibacillary (PB) leprosy case had tour specimens taken, all of which were negative for both tests in both laboratories. Nine of the 16 smear-positive tuberculosis cases were confirmed by culture.
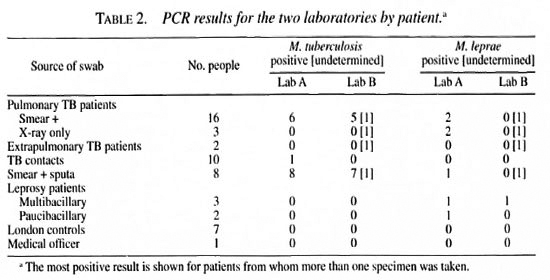
Thirty patients contributed duplicate swabs for each laboratory (including pairs from each of the leprosy patients). Excluding those with undetermined results. Laboratory A found agreement in 25/29 pairs (86%) for M. tuberculosis, and in 24/29 (83%) for M. leprae. Laboratory B found agreement for all 20 pairs for both species of mycobacteria when both results were determined, but only two of the tuberculosis patients with positive results had paired results, and the only positive leprosy results came from the MB patient with six specimens.
Both laboratories detected M. tuberculosis in all of the (positive control) smear-positive sputa (although Laboratory B was unable to obtain results from one of these swabs because of inhibitors); neither laboratory found any positive results among the London (negative) controls.
Laboratory A detected M. tuberculosis by PCR in 8/32 (25%) specimens from pulmonary cases and in 1 of 13 (8%) from tuberculosis contacts (one not determined due to the presence of inhibitors); whereas Laboratory B identified M. tuberculosis in 7/33 (21%) specimens from pulmonary tuberculosis cases (seven not determined) and in none of the 13 specimens from tuberculosis contacts (one not determined). Among the pulmonary tuberculosis cases detected, three were detected by both laboratories.
M. leprae were found by each laboratory in all six nasal swabs obtained from one MB (slit-skin smear-positive) leprosy patient. Of the other 4 leprosy patients (2 MB and 2 PB), only one PB patient had a positive specimen (Laboratory A). No household contacts of leprosy patients were tested, but M. leprae were reported in nasal swabs from 4 of 21 tuberculosis patients and one tuberculosis sputum by Laboratory A, but not by Laboratory B.
DISCUSSION
Since PCR techniques are very sensitive and capable of delecting even nonviable bacilli, and since smear-positive tuberculosis patients may produce 105 to 107 bacilli per ml of sputum, some of which get into airborne droplets which can be exhaled or inhaled, we had expected to detect M. tuberculosis in the noses of the majority of these patients. In practice, bacilli were detected in about one third of the smear-positive patients and in only one specimen from the contacts of tuberculosis patients.
The techniques used by Laboratory B have been shown to be able to detect one leprosy bacillus or 5-20 tuberculosis bacilli using purified genomic DNA (4,12). Equivalent figures for Laboratory A are 20 M. leprae and 2 tuberculosis bacilli (3,7). In general, however, larger numbers of organisms appear to be necessary to yield a positive result by PCR when using clinical samples (1,6). It is possible that some degradation of nucleic acid occurred during the transport from Malawi, although efforts were made to keep the specimens cold. Among the sputum specimens, in which larger numbers of bacilli were present, sensitivity was 100%.
Where the results from the two laboratories differed, it is difficult to know whether this reflects lack of sensitivity by one or of specificity by the other. M. leprae have previously been identified in the noses of persons without leprosy from endemic areas (5) and the "failure" of Laboratory B to detect M. leprae in any specimens except those from one MB leprosy patient may be attributable to the lack of a purification step in this laboratory when inhibitors were found: of the eight nasal swabs from the four tuberculosis patients found positive for M. leprae by Laboratory A, Laboratory B could only determine results for three. Alternatively, the five specimens from tuberculosis patients found positive for M. leprae by Laboratory A may be false-positives. This is unlikely to be due to crossreaction since, if crossreaction did occur, it would be expected preferentially in the sputum specimens since they contain the most M. tuberculosis , and this was not the case. Laboratory contamination is a possible explanation for these results, and could have occurred tit the DNA purification step since all five of these swabs showed inhibition initially and underwent purification during M. leprae tests (compared to 26 specimens overall).
These results show that M. tuberculosis can be found in the nose of some pulmonary tuberculosis patients, albeit in fewer than might be expected. The results from the positive and negative controls support the accuracy of the results of the assays, but the comparisons with M. leprae and between laboratories suggest that the technique is less reliable and probably less sensitive than might be hoped. Given the poor yield of positive PCR results from nasal swabs from tuberculosis cases, the finding of only one positive result among their contacts is not surprising. The results to date come only from a small number of patients, and it would be useful to expand this investigation as an adjunct to studies of transmission of mycobacterial infections in different settings.
Acknowledgments. The Karonga Prevention Study is funded primarily by LEPRA (British Leprosy Relief Association) and ILEP (International Federation of Anti-Leprosy Organizations), with contributions from the WHO/UNDP/World Bank Special Programme for Research and Training in Tropical Diseases and the Tuberculosis Division of WHO. SJ was supported by LEPRA, and this investigation received financial support from the Netherlands Leprosy Relief Association. The authors thank the Government of the Republic of Malawi for their interest in and support of the project, and the Malawi Ministry of Health for permission to publish the paper.
REFERENCES
1. CLARRIDGE, J. E., III. SHAWAR, R. M., SCHINNICK, T. M. and PLIKAYTIS, B, B. Large-scale use of polymerase chain reaction for detection of Mycobacterium tuberculosis in a routine mycobacteriology laboratory. J. Clin. Microbiol. 31(1993)2049-2056.
2. DE KANTOR, I. N. and ROSWURM, J. D. Mycobacteria isolated from nasal secretions of tuberculin test reactor cattle. Am. J. Vet. Res. 39(1978)1233-1234.
3. DE WIT, M. Y. L., DOUGLAS, J. T., MCFADDEN, J. and KLATSER, P. R. Polymerase chain reaction for detection of Mycobacterium leprae in nasal swab specimens. J. Clin. Microbiol. 31(1993)502-506.
4. JAMIL, S., WILSON, S. M., HACKETT, M., HUSSAIN, R. and STOKER, N. G. A colorimetric PCR method for the detection of Mycobacterium leprae in skin biopsies from leprosy patients. Int. J. Lepr. 62(1994)512-520.
5. KLATSER, P. R., VAN BEERS, S., MADJID, B., DAY,R. and DeWIT, M. Y. L. Detection of Mycobacterium leprae nasal carriers in populations for which leprosy is endemic. J. Clin. Microbiol. 31(1993)2947-2951.
6. KOLK, A. H. J., SCHUITEMA, A. R. J., KUIJPER, S., VAN LEEUWEN, J., HERMANS, P. W. M., VAN EMBDEN, J. D. A. and HARTSKEERL, R. A. Detection of Mycobacterium tuberculosis in clinical samples by using polymerase chain reaction and a non-radioactive detection system. J. Clin. Microbiol. 30(1992)2567-2575.
7. Kox, L. F. F., RHIENTHONG, D., MEDO MIRANDA, A., UDOMSANTLSUK, N., ELLIS, K., VAN LEEUWEN, J., VAN HEUSDEN, S., KUIJPER, S. and KOLK, A. H. J. A more reliable PCR for detection of Mycobacterium tuberculosis in clinical samples. J. Clin. Microbiol. 32(1994)672-678.
8. McILROY, S. G., NEILL, S. D. and MCCRACKEN, R. M. Pulmonary lesions and Mycobacterium bovis excretion from the respiratory tract of tuberculin reacting cattle. Vet. Rec. 118(1986)718-721.
9. PATTYN, S. R., URSI, D., LEVEN, M., GRILLONE, S. and RAES, V. Detection of Mycobacterium leprae by the polymerase chain reaction in nasal swabs of leprosy patients and their contacts. Int. J. Lepr. 61(1993)389-393.
10. RAO, S., RAU, P. V, SAHOO, R. C, RAJAN, R. and MURTHY, P. S. Primary nasal tuberculosis. Letter Tuber. Lung Dis. 73(1992)305.
11. SWART, J. G., DE FLAMINGH, D. Q. and HAMERSMA,T. Histologically detected extrapulmonary tuberculosis in the head and neck region; a review of 222 cases. S. Afr. Med. J. 71(1987)700-702.
12. WILSON, S. M., MCNERNEY, R., NYE, P. M., GODFREY-FAUSSETT, P. D., STOKER, N. G. and VOLLER, A. Progress toward a simplified polymerase chain reaction and its application to diagnosis of tuberculosis. J. Clin. Microbiol. 31(1993)776-782.
1. M.D., Karonga Prevention Study, P. O. Box 46, Chilumba, Malawi.
2. M.R.C.P., London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT, U.K.
3. Ph.D., London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT, U.K.
4. Ph.D., London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT, U.K.
5. Ing., Royal Tropical Institute, Meibergdreef 39, 1105 AZ Amsterdam, The Netherlands.
6. Karonga Prevention Study, P. O. Box 46, Chilumba, Malawi.
7. Ph.D., London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT, U.K.
8. Ph.D., Royal Tropical Institute, Meibergdreef 39, 1105 AZ Amsterdam, The Netherlands.
Reprint requests to Dr. J.R. Glynn at the above address or FAX 44-171-436-4330.
Present address for Dr. S. Jamil; Department of Microbiology, The Aga Khan University, P. O. Box 3500, Karachi, Pakistan.
Received for publication on 9 February 1996.
Accepted for publication in revised form on 21 June 1996.