- Volume 63 , Number 2
- Page: 241–8
Variants of HLA-DR2/DR51 group haplotypes and susceptibility to tuberculoid leprosy and pulmonary tuberculosis in Asian Indians
ABSTRACT
This study reports our observations on the correlation between HLA-DR2 subtypes and their DR-DQ haplotypes in patients with tuberculoid (TT) leprosy and pulmonary tuberculosis (PTB). DRB 1*1501 was significantly increased in patients with PTB (90%) as compared to controls (p < 0.05); whereas the prevalence of DRB 1*1502 was significantly increased in patients with TT leprosy (p < 0.05), suggesting allele-specific binding of the pathogen to form disease-causing motifs to the T-cell receptor. Among DR2-DQ haplotypes, the deviation was noted in the distribution of unique and common haplotypes in patients with TT leprosy and PTB. A significant decrease of haplotype DRB1*1501-DRB5*0101-DQA1*0102-DQB 1*0502 in TT leprosy and a significant increase of DRB 1 * 1 501-DRB5*0101DQA1*0103-DQB 1*0601 in PTB patients were observed. The occurrence of specific DR2 subtypes and their haplotypes in the two disease groups suggests their involvement in disease pathogenesis.RÉSUMÉ
Cette étude rapporte nos observations sur la correlation entre les sous-types HLA-DR2 et leurs haplotypes DR-DQ chez les patients présentant une lèpre tuberculoide (TT) et une tuberculose pulmonaire (TBP). Le DRB1*1501 était significativement élevé chez les patients avec TBP (90%), comparés aux témoins (p < 0.05); tandis que la prevalence du DRB1*1502 était significativement élevée chez, les patients avec une lèpre TT (p < 0.05), suggérant un lien, spécifique pour Palíele, du pathogène aux éléments responsables de la forme de la maladie sur le récepteur de la cellule-T. Parmi les haplotypes DR2-DQ, on a noté une déviation dans la distribution d'haplotypes uniques et communs chez les patients avec une lèpre TT et une TBP. Une diminution significative de l'haplotype DRB1*I501DRB5*0101-DQA1*0102-DQB1*0502 dans la lèpre tuberculoide et une augmentation significative de DRB1 * 1501 -DRB5*0501 -DQA 1 *0103-DQB1 *0601 chez, les patients TBP ont été observées. La présence de sous-types DR2 spécifiques et de leurs haplotypes dans les deux maldics suggère leur implication dans la pathogénèse.RESUMEN
En este estudio reportamos nuestras observaciones sobre la correlación entre los subtipos de HLA-DR2 y sus haplotipos DR-DQ, en pacientes con lepra tuberculide (TT) y en pacientes con tuberculosis pulmonar (PTB). Comparando con los controles sanos, DRB1 * 1501 estuvo significativamente incrementado en los pacientes con PTB (90%) (p < 0.05) mientras que DRB1 * 1502 estuvo significativamente más elevado en los pacientes con lepra TT (p < 0.05), sugiriendo un cnlazamiento alelo-específico del patógeno con alguna secuencia particular del receptor de la célula T. Entre los haplotipos DR2-DQ, se notó la desviación en la distribución de haplotipos únicos y comunes en los pacientes con lepra TT y PTB. Se observó una disminución significativa del haplotipo DRB1-1 * 501-DRB5*0101 -DQA1 *0102-DQB 1 *0502 en los pacientes con lepra TT y un incremento significativo de DRB1*1501 * DRB5 * 0101-DQA1 * 0103-DQB1 * 0601 en los pacientes con PTB. La ocurrencia de subtipos específicos de DR2 y sus haplotipos en las dos enfermedades, sugiere su participación en la patogénesis de las enfermedades.Although the immunobiology ofleprosy/ plumonary tuberculosis (PTB) has received much attention for several years, the individual differences in resistance and response to the bacilli are still unknown. The severity of these infectious diseases is associated with T-cell immune responsiveness against Mycobacterium leprae and M. tuberculosis rather than direct damage by the bacilli (2,10). Since HLA class II molecules have been demonstrated as products of the immune response (Ir) and/or immune suppression (Is) genes (1,18), extensive polymorphism in these genes may lead to differences in susceptibility to infection and/or expression of disease.
We have demonstrated earlier a strong association between HLA-DR2 and Asian Indian patients with tuberculoid (TT) leprosy (8,14,21) and PTB (19, 20). With the introduction of the molecular techniques of HLA typing, DR2 has been split into several subtypes, the genes of which are expressed in two DRB loci: DRB1 and DRB5; together they constitute the DR51 group of haplotypes. Using polymerase chain reaction (PCR) amplified DNA products and allele-specific oligonucleotide probes, five DR2B1 alleles (1501, 1502, 1503, 1601, 1602) and four DRB5 alleles (0101, 0102, 0201,0202) have been defined recently (3,l6). In addition, heterogeneity in DR2 haplotypes results from associated alleles of the DQA and DQB genes (15). The distribution of the subtypes of DR2 in tuberculoid leprosy and PTB is, however, not yet known. The objectives of the present study, therefore, were to investigate the occurrence of various DR2 subtypes and their polymorphic DQA 1-DQB 1-associated genotypes in TT leprosy and PTB and their comparison with healthy controls.
MATERIALS AND METHODS
Study populations. A total of 85 unrelated patients with TT leprosy (average age 37 years, 47 males and 38 females), 45 patients with PTB (average age 35 years, 26 males and 19 females), and 104 healthy controls (average age 38 years, 59 males and 45 females) were included in this study. All patients and controls represented a fairly homogeneous ethnic group of north Indian Hindus belonging to the states of Punjab, Haryana, Uttar Pradesh and Delhi. Leprosy patients were classified according to the fivegroup system of Ridley and Jopling (17) based on clinical examination, skin-smear bacteriology and histopathology of the skin-lesion biopsy material. Active PTB was diagnosed by the presence of acid-fast bacilli (AFB) in direct sputum smears or in cultures, and by the standard clinical and radiological investigations. Sputum smears were examined at least three times in each case for AFB by auraminc stain fluorescent microscopy (9). All patients were obtained from the All India Institute of Medical Sciences and Lala Ram Sarup Institute for TB and Allied Diseases, Mehrauli, New Delhi. A qualified Leprologist, a bacteriologist and an histopathologist were involved in the diagnosis.
The controls consisted of healthy volunteers and laboratory personnel derived from the same ethnic and socioeconomic status who had no family history or symptoms of leprosy, tuberculosis or other related diseases. These were age- and sexmatched with the patients.
Families. The assignment of HLA-DR2/ DQ1 haplotypes was reconfirmed by segregation analysis of parental HLA haplotypes in families. A total of 30 families (consisting of 120 individuals) tested for this purpose included 7 with leprosy, 2 with PTB, and 20 healthy families. Due to the small number of lepromatous leprosy cases available in these families, data on these were not included for statistical analysis.
HLA typing. Twenty ml of heparinized blood (20 IU of preservative-free heparinc/ ml) was freshly drawn from each individual and lymphocytes were separated on a Ficoll-Hypaque density gradient. T and B cells were separated using nylon wool columns and HLA class I (HLA-A, -B, -C) and class II (DR and DQ) phenotypes were determined for all patients and healthy subjects using the standard two-stage NIH microlymphocytotoxicity assay (23). All DR2positive individuals thus identified were further analyzed by the molecular typing techniques of PCR using sequence specific oligonucleotide probes (SSOPs) to determine their subtypes. In this way, 39 unrelated patients with tuberculoid leprosy, 20 with pulmonary tuberculosis, and 46 healthy controls were analyzed by DNA typing.
Extraction of cellular DNA. DNA was prepared from peripheral blood leukocytes according to the method previously described (16). Quantification of the DNA was done by checking the optical density (OD) at wave lengths 260 nm and 280 nm. All samples had their 260/280 OD ratios between 1.7 and 2.0.
PCR amplification of DR and DQ genes. DNA samples were PCR amplified using a programmable thermocycler (Perkin Elmer-Cetus) and different sets of specific primers for the polymorphic region (second exons) of the DRB, DQA and DQB genes as described earlier (16). A set of group- and allele-specific primers were used for the amplification of DR2B1 and DR2B5 genes. The PCR reaction mixture consisted of 0.1-0.5 mg of genomic DNA; 200 μM each of dATP, dCTP, dGTP and dTTP; 20 pmol of each of the primers; 5 units of Taq DNA polymerase, as well as a buffer containing 50 mM KC1, 10 raM Tris-HCl (pH 8.4), 4 mM MgC12, 60 μg/mg bovine serum albumin, and double distilled water up to a final volume of 100μl. The PCR mixture was then covered with 100 μl of mineral oil. The reaction mixture was subjected to 40 temperature cycles of 30 sec at 97ºC allowing the DNA to denature, 30 sec at 55ºC for primers to anneal, and 30 seconds at 72ºC for annealed primers to extend. Finally, there was an additional 10-min incubation at 74ºC to complete DNA synthesis. Five percent of the PCR product was electrophoresed in 2% agarose in TBE buffer (0.045 M Trisborate, 0.001 M EDTA) to check for the efficiency and specificity of the reaction. DNA from the 9th and 10th International Histocompatibility Workshop (IHW) cell lines as well as from selected local panel cells representing the known specificities of DRB, DQA and DQB served as positive and negative controls.
Biotinylated oligonucleotide probes. The sequences of the oligonucleotide probes used in this study are based on those employed in the 11th IHW (11). These were synthesized using cyanocthyl phosphoramidite chemistry in a cyclone DNA synthesizer (Milligen/Millipore) and identified known alleles at the DRB1, DRB3, DRB4, DRB5, DQA1 and DQB1 loci. All oligonucleotide probes were biotinylated at the 5' end and were designed as 20 and 18 mers, respectively, for DR and DQ typing. Special attention was given to probes that could detect HLA-DR2 subtypes in both the DRB1 and DRB5 genes.
Dot blot hybridization. Of each amplified DNA sample, 5 μl was denatured with 100 μl 0.4N NaOH, 10 mM EDTA for 10 min at room temperature and dot blotted onto prewetted Hybond N+ membranes (Amersham, U.K.) using Bio-Dot microfiltration apparatus (Biorad). After washing five times with standard saline phosphatebuffered EDTA (SSPE), the DNA was crosslinked to the membranes by UV-irradiation using a stratalinker 1800 (Stratagene, USA) set at 25 mj. Blots were prehybridized in tetramethylammonium chloride (TMAC1) solution consisting of 3 M TMAC1, 5 mM EDTA, 50 mM Tris (pH 7.5), 1% SDS and 100 μg/ml boiled herring sperm DNA for a period of ½ - 2 hr at varying temperatures (55ºC for DRB, 50ºC for DQA and DQB). Biotinylated SSOPs (1 pmol/ml) were added to the solution and hybridization continued for 1-4 hr (sometimes overnight) at 50ºC for DQ probes or at 55ºC for DR probes. Membranes were then washed twice in washing buffer (1 x SSPE, 0.1% SDS) at room temperature. Following this, they were subjected to a "critical" wash step in TMAC1 buffer at varying temperatures between 56ºC-65ºC (depending on the SSOP used) for 15 min. They were then washed twice in the washing buffer at room temperature and processed further for final detection.
Membranes were incubated in Streptavidin-HRP (10 ml washing buffer + 1 μl stock solution, 2 mg/ml) for 15-20 min. Generally 4-8 membranes were incubated in 50 ml solution. The membranes were washed twice in washing buffer for 5 min each followed by another wash step in blocking buffer (1 M urea, 0.1 M NaCl, 5% Triton X-100, 1% dextran sulfate) for 5 min. The membranes were again washed in washing buffer once and were now ready for the enhanced chemiluminescence (ECL) detection system using the Amersham (U.K.) ECL kit. For this purpose, 10 ml solution 1 and 10 ml solution 2 were mixed together and two membranes at a time were soaked in the mixture for 1-1 ½ min. Membranes were blotted dry and exposed to Kodak X-Omat film for 10-60 sec.
Statistical analysis. Distribution of subtypes of DR2 and DQ 1 specificities was performed by the χ2 test and the probability (p) value was obtained to ascertain the significance. The p values were corrected by multiplication with the number of alleles studied in each locus (24). Relative risk (RR) values were calculated by a modified method of Woolf and Haldane (22).
RESULTS
Distribution of HLA-DR2 subtypes. The distribution of DRB1, DQA1 and DQB1 alleles in DR2-positive patients with TT leprosy and PTB was compared with that in normal healthy controls (The Table). DRB1 * 1501 and DRB1 * 1502 were the most frequent alleles in DR2-positive subjects in all three groups. No examples of DRB 1 * 1503 and DRB 1*1601 were encountered. The prevalence ofDRBl*1501 in patients with PTB (90%) was significantly higher than that in normal healthy controls (65.2%, p < 0.05, RR = 5.28) and in patients with TT leprosy (51.3%, p < 0.005). This allele, however, did not reveal any deviation in TT leprosy as compared to controls. On the other hand, DRB1*1502 occurred significantly more frequently in TT leprosy (64.1 %) as compared to healthy controls (39.1%, p < 0.025, RR = 2.73) and that in patients with PTB (15%, p < 0.001). This allele was significantly lower in patients with PTB as compared to controls (p < 0.05, RR = 0.25).
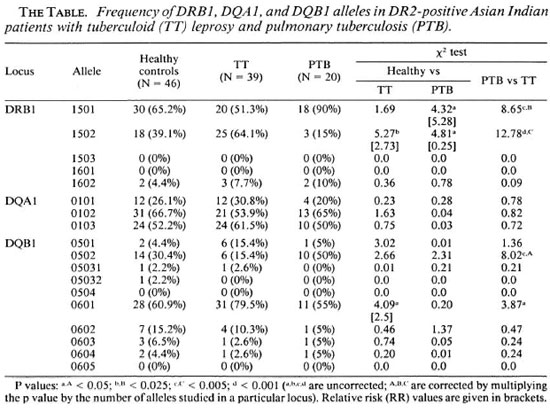
No differences were observed in the distribution of DQA1 alleles in the two groups of patients and healthy controls. Of the DQB1 alleles, the frequency of DQB1 *0601 was higher in TT leprosy (79.5%) compared with normal controls (60.9%, p < 0.05, RR = 2.5) and patients with PTB (55%, p < 0.05). Similarly, the frequency of DQB 1*0502 was higher in PTB patients (50%) compared to normal controls (30.4%), although this difference was not statistically significant. However, the difference in the prevalence of DQB 1*0502 in the two groups of patients was statistically significant (p < 0.005).
DR2-associated DRB1-DRB5-DQA1-DQB1 genotypes. Next, we compared the distribution of DRB1-DRB5-DQA1 -DQB 1 genotypes in the three groups of subjects. A total of nine different DR2-associated haplotypes were encountered (Fig. 1). Two of these nine haplotypes (nos. 3 and 4) showed deviation in their frequency distribution in the patient group as compared to controls. Haplotype no. 3 (DRB 1*1501 -DRB5*0101DQA1*0102-DQB 1*0502) was significantly decreased in patients with TT leprosy (7.7%) compared to controls (23.9%, p < 0.05, RR = 0.22). On the other hand, haplotype no. 4 (DRB1*1501-DRB5*0101DQA1*0103-DQB 1*0601) occurred with significantly increased frequency in patients with PTB (35%) as compared to healthy controls (13%, p < 0.05, RR = 3.5).
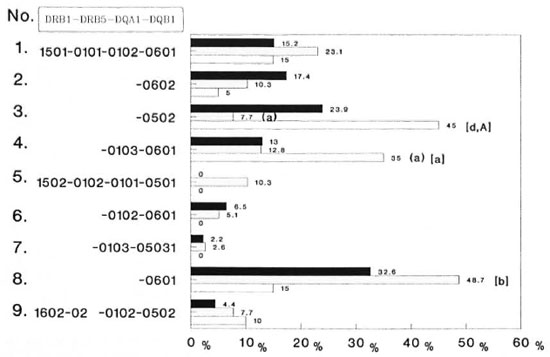
Fig. 1. Frequency of DR2-DQ1 genotypes in Asian Indian patients with tuberculoid leprosy (TT) and pulmonary tuberculosis (PTB); p values = A, A < 0.05; B < 0.025; D < 0.001 (A, B, D are uncorrected; A is corrected by multiplying the p value of the number of alleles studied in a particular locus); p values presented in parentheses are healthy vs diseased; those presented in brackets refer to TT leprosy vs PTB. ■ = healthy (N = 46);  = TT leprosy (N = 39); □ = PTB (N = 20).
= TT leprosy (N = 39); □ = PTB (N = 20).
When an analysis was made among diseased patients, both of these haplotypes were increased significantly in PTB as compared to patients with TT leprosy (haplotype 3, p < 0.001; haplotype 4, p < 0.05). The frequency of haplotype no. 8 (DRB1*1502DRB5*0102-DQA1*0103-DQB1*0601) was decreased significantly in patients with PTB (15%) as compared to those with TT leprosy (48.7%, p < 0.025). Haplotype no. 5 (DRB1 * 1502-DRB5*0102-DQA 1*0101-DQB 1*0501) was encountered only in patients with TT leprosy (10.3%) while haplotypes 6 and 7 were totally absent in PTB.
Leprosy families. The pedigrees of all seven families with leprosy are given in Figure 2. All families were selected at random with no bias toward their DR2 status and had at least two affected members (multicase families). A total of 16 patients with TT leprosy and 6 with lepromatous leprosy were available in these families. The most frequent DR2-positive haplotype encountered is haplotype no. 8 (DRB1*1502DRB5*0102-DQA 1*0103-DQB 1*0601). Except for families nos. 2 and 7, DR6-associated haplotypes were absent in all seven families.
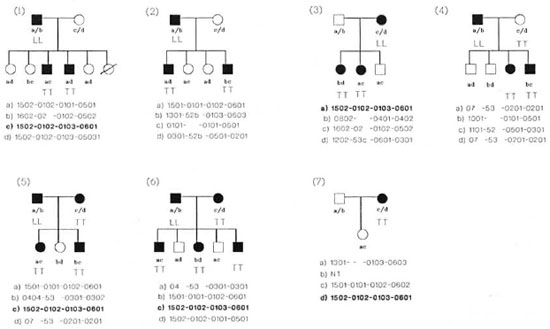
Fig. 2. Pedigree charts of the seven families studied: a, b, c, and d indicate HLA genotypes; haplotype in bold letters indicates the most common haplotype in TT leprosy patients. LL = lepromatous leprosy; TT = tuberculoid leprosy; NT = not tested for HLA.
DISCUSSION
We have reported earlier that TT leprosy and PTB are associated with DR2 (14). The results in our present study extend those data and further demonstrate that different subtypes of DR2 and DR2-DQ1 haplotypes are associated with TT leprosy and PTB as compared to normal controls. In addition, some alleles showed a significant preference in their prevalence in these two groups of patients. For example, the frequency of DRB 1*1501 was significantly increased in patients with PTB; whereas the prevalence of DRB 1 * 1502 was strongly associated with TT leprosy. Sequence analyses have disclosed that only one amino acid variation can discriminate the products of DRB1*1501 from DRB1*1502 and DRB 1*1601 from DRB1*1602 (l3). In particular DRB1*1501 carries valine at amino acid position 86 while it is substituted with glycine in DRB 1*1502. DRB1*1601 has the aromatic amino acid phenylalanine at position 67 which is substituted by aliphatic leucine in DRB 1*1601. The three-dimensional crystallographic structure of the human class II HLA molecule (4) has revealed that the positions of residues 67 and 86 are at the alpha helix of the beta chain facing the antigen-binding groove and are actively involved in binding a foreign peptide. Accordingly, the peptide binding and subsequent immune triggering depends critically on these single amino acid variants. It is possible that the DRBP1501 and DRB1*1502 alleles may be selectively implicated in the presentation of pathogenic peptides of my cobacteria, leading to the development of TT leprosy and PTB. In this model, the putative disease influencing the MHC allele-pathogen motif will be shown to the T-cell receptor leading to a detrimental cellular immune response responsible for the disease.
Haplotype no. 3 appears to be a common one encountered in the Indian population (23.9%). It has only been reported among Javanese (10.3%) and south Chinese (6%), and has a very low frequency (0.8%) in Melanesians (7). This haplotype was most commonly encountered in patients with PTB. A recent WHO survey indicates that PTB prevalence is most common in developing nations, including India (12). Incidentally, another haplotype (no. 4) which appeared significantly raised in patients with PTB is unique to the Indian population, and has not been reported in any other ethnic group so far (15). A significant increase of these haplotypes in patients with PTB may suggest that in addition to nutritional and environmental factors, the existence of such unique haplotypes in Indians could have contributed to the hyperendemicity of PTB. However, another unique haplotype of Asian Indians, DRB1*1502-DRB5*0102DQA1*0103-DQB 1*05031 (haplotype no. 7), was not encountered in PTB.
Out of the two common haplotypes of DR2 that occur worldwide, only one (haplotype no. 8) occurred with an appreciably increased frequency (48.7%) in TT leprosy, while the other haplotype (no. 2) is within the normal range (10.3%). Haplotype no. 8 occurs with moderate frequency in North American Caucasoids (9.4%) and in Latin Americans (7.6%) (6), while it is the most predominant haplotype among the north Chinese (34.2%) (7) and north Indians (32.6%). At this stage, it is premature to say whether such a distribution of this particular haplotype might have contributed toward the increased incidence of leprosy in India and other developing countries. Further, the roles of environmental, nutritional and other non-MHC genetic factors, such as the one recently described for governing susceptibility to M. leprae infections (21), are of paramount importance. Further studies in other ethnic groups involving sporadic patients with leprosy and tuberculosis is desirable.
Out of the seven families with leprosy in this study, five were found to carry haplotype no. 8 (Fig. 2). Further, haplotype no. 5, DRB1*1502-DRB5*0102-DQA1*0101-DQB 1*0501, was found only in TT leprosy patients and not in healthy and PTB groups. This haplotype occurs predominantly among Javanese (7) and among the Thai population (8).
This study provides evidence for the distribution of different subtypes of DR2 and their unique and common DR-DQ haplotypes in TT leprosy and PTB. It is likely that peptides originating from M. leprae and M. tuberculosis bind preferentially to HLA-DR2 allelic forms characterized by their molecular subtypes and/or haplotypes and stimulate T-cell clones that result in detrimental immune responses in these two mycobacterial diseases. In this context, delineation of relevant peptides is important, particularly in understanding the immunopathogenesis of lepromatous leprosy.
Acknowledgment. We thank Prof. D. P. Singal, Mc-Master University, Ontario, Canada, for his critical comments. This work was supported by grants from the Department of Biotechnology, Ministry of Science and Technology, Government of India; the National Reference Center for Histocompatibility Testing, Leiden, The Netherlands, and the Macropa Foundation, Leiden, The Netherlands.
REFERENCES
1. BENACERRAF, B. Role of MHC gene products in immune regulation. Science 212(1981)1229-1238.
2. BLOOM, B. R. and MEHRA, V. Immunological unresponsiveness in leprosy. Immunol Re v. 80(1984)5-28.
3. BODMER, J. G., MARSH. S. G. E., ALBERT, E. D., BODMER, W. F., DUPONT, B., ERLICH, H. A., MACH, B., MAYR, W. R., PARHAM, P., SASAZUKI, T., SCHREUDER, G. M. T., STROMINGER, J. L., SVEJGAARD, A. and TERASAKI, P. I. Nomenclature for factors of the HLA system, 1991. Tissue Antigen 39(1992)161-173.
4. BROWN, J. H., JARDETZKY, T. S., GORGA, J. C, STERN, L. J., URBAN, R. G., STROMINGER, J. L. and WILEY, D. C. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364(1993)33-39.
5. DE VRIES, R. R. P., MEHRA. N. K., VAIDYA, M. C, GUPTE, M. D., KHAN. P. M. and VA N ROOD, J. J. HLA linked control of susceptibility to tuberculoid leprosy and association with HLA-DR types. Tissue Antigens 16(1980)294-304.
6. FERNANDEZ-VINA, M. A., GAO, X. J., MORAES, M. E., MORAES, J. R., SALATIEL, I., MILLER, S., TSAI, J., SUN, Y. P., AN, J. B., LAYRISSE, Z., GAZIT, E., BRAUTBAR, C. and STASTNY, P. Alleles at four HLA class II loci determined by oligonucleotide hybridization and their association in five ethnic groups. Immunogenctics 34(1991)299-312.
7. GAO, X. J. and SERJEANTSON, S. W. Heterogeneity in HLA-DR2 related DR, DQ haplotypes in eight populations of Asia Oceania. Immunogenctics 34(1991)401-408.
8. GIPHART, M. J., D'AMARO, J., DRABBELS, J., SCHIP PER, R., TANEJA, V., MEHRA, N. K., SCHREUDER, G. M. T. and VERDUIJN, W. Analysis of HLA-DR1 and DR2 alleles. In: HLA 1991. Tsuji, K., Aizawa, M. and Sasazuki, T., eds. Oxford: Oxford University Press, 1992, pp. 457-461.
9. HOLST, E., MITCHISON, D. A. and RADHAKRISHNA, S. Examination of smears for tubercle bacilli by fluorescence microscopy. Indian J. Med. Res. 47(1959)495-496.
10. KAUFMANN, S. H. E. Tuberculosis: the role of the immune response. Immunologist 1(1993)109-114.
11. KIMURA, A. and SASAZUKI, T. Eleventh International Histocompatibility Workshop reference protocol for the HLA DNA-typing technique. In: HLsi 1991. Tsuji, K.., Aizawa, M. and Sasazuki, T., eds. Oxford: Oxford University Press, 1992, pp. 397-419.
12. KOCHI, A. The global tuberculosis situation and the new control strategy of the World Health Organization. Tubercle 72(1991)1-6.
13. MARSH, S. G. E. and BODMER, J. G. HLA class II nucleotide sequences, 1992. Immunogenetics 37(1993)79-94.
14. MEHRA, N. K. Role of HLA linked factors ingoverning susceptibility to leprosy and tubercu-losis. Trop. Med. Parasitol. 41(1990)352-353.
15. MEHRA, N. K., BOUWENS, A. G. M., NAIPAL, A.,RAJALINGAM, R., GRUBIC, Z., TANEJA, V., TILANUS, M. G. J. and GIPHART, M. J. Asian Indian HLA-DR2, DR4 and DR52 related DR-DQ genotypes analyzed by PCR based non-radioactive oligonucleotide typing: unique haplotypes and anovel DR4 subtype. Hum. Immunol. 39(1994)202-210.
16. MEHRA, N. K., VERDUIJN, W., TANEJA, V., DRAB-BELS, J., SINGH, S. P. N. and GIPHART, M. J. Analysis of HLA-DR2 associated polymorphism byoligonucleotide hybridization in an Asian Indianpopulation. Hum. Immunol. 32(1991)246-253.
17. RIDLEY, D. S. and JOPLING, W. H. Classificationof leprosy according to immunity; a five-groupsystem. Int. J. Lepr. 34(1969)255-273.
18. SCHWARTZ, R. H. Immune response (Ir) genes ofthe murine major histocompatibility complex.Adv. Immunol. 38(1986)31-201.
19. SINGH, S. P. N., MEHRA, N. K., DINGLEY, H. B.,PANDE, J. N. and VAIDYA, M. C. HLA-A, -B, -C, and -Dr antigen profile in pulmonary tuberculosisin north India. Tissue Antigens 21(1983)380-384.
20. SINGH, S. P. N., MEHRA, N. K., DINGLEY, H. B.,PANDE, J. N. and VAIDYA, M. C. Human leukocyte antigen (HLA)-linked control of susceptibilityto pulmonary tuberculosis and association withHLA-DR types. J. Infect. Dis. 148(1983)676-681.
21. SKAMENE, E. Genetic control of susceptibility tomycobacterial infections. Rev. Infect. Dis. Suppl. 2(1989)S394-5399.
22. SVF_JGAARD A., PLATZ, P. and RYDER, L. P. HLA and disease 1982: a survey. Immunol. Rev. 70(1983) 193-201.
23. TERASAKI, P. I. and MCCLELLAND, J. D. Microdroplet assay of human serum cytotoxin. Nature 204(1964) 998-999.
24. TIWARI, J. L. and TERASAKI, P. I. HLA and Disease Association . New York: Springer Verlag, 1985,pp. 18-27.
25. VAN EDEN, W., DE VRIES, R. R. P., MEHRA, N. K.,VAIDYA, M. C., D'AMARO, J. and VAN ROOD, J. J. HLA segregation of tuberculoid leprosy: confirmation of the DR2 marker. J. Infect. Dis. 141(1980)693-701.
1. Ph.D.;Department of Histocompatibility and Immunogenctics, All India Institute of Medical Sciences, New Delhi 110029, India.
2. M.Phil.; Department of Histocompatibility and Immunogenctics, All India Institute of Medical Sciences, New Delhi 110029, India.
3. Ph.D., Department of Immunohematology and Blood Bank, University Hospital, Leiden, The Netherlands.
Received for publication on 22 April 1994.
Accepted for publication in revised form on 21 December 1994.