- Volume 63 , Number 1
- Page: 42–7
Application of polymerase chain reaction for the detection of Mycobacterium leprae DNA in specimens f rom treated leprosy patients
ABSTRACT
In this study of leprosy patients apparently cured by dapsonc monotherapy, the polymerase chain reaction (PCR), one of the most reliable and sensitive DNA-bascd assays, was used for the specific detection of Mycobacterium leprae DNA. Sputum and slit-skin samples f rom 44 such patients at Baba Baghi Leprosy Sanatorium in Iran were examined. Primers for a 530-base-pair fragment of the gene encoding the 36-kDa antigen of M. leprae were used for the study. The PCR results were compared with microscopy for acid-fast bacilli. Of the 44 sputum samples, 2 were positive by PCR (4.5%) and of the 44 slit-skin swabs taken f rom the same patients, 10 were PCR positive (22.7%). Only one patient was PCR positive for both sputum and slit-skin specimens (2.3%). No positive results were found by acid-fast microscopy. In total, 11 of44 (25%) patients in this study were found to be PCR positive for M. leprae, and it was thought probable that this indicated the presence of live organisms. Particularly interesting was the statistically significant association of positive results f rom slit-skin swabs with paucibacillary rather than multibacillary leprosy. It is suggested that whereas relapse or immunological reaction in paucibacillary disease may result f rom surviving organisms, in multibacillary leprosy this may be due to re-infection.RÉSUMÉ
Dans cette étude de malades de la lèpre apparemment guéris par une monothérapie à la dapsonc, la réaction de polymerase en chaîne (PCR), l'un des tests les plus fiables et les plus sensibles basés sur l'ADN, fut utilisée pour la détection spécifique d'ADN de Mycobacterium leprae. Des expectorations et des frottis cutanés de 44 de ces patients du Sanatorium Baba Baghi pour la lèpre ont été examinés. Des amorces pour un fragment pairé de 530 bases du gène encodant l'antigène de 36 kDa de M. leprae ont été utilisées pour l'étude. Les résultats de PCR ont été comparés avec la microscopic pour la recherche de bacilles acido-résistants. Parmi les 44 échantillons de crachats, 2 étaient positifs par PCR (4.5%) et des 44 frottis cutanés prélevés chez les mêmes patients, 10 étaient positifs à la PCR (22.7%). Seul un patient était positif pour les expectorations et les frottis cutanés (2.3%). Aucun résultat positif n'a été trouvé par microscopic Au total, 11 des 44 patients (25%) de cette étude ont été trouvés positifs à la PCR pour M. leprae, et il a semblé probable que cela indiquait la présence d'organismes vivants. D'un intérêt particulier était l'association statistiquement significative de résultats positifs à partir de frottis cutanés, pour la lèpre paucibacillairc plutôt que pour la lèpre multibacillaire. L'hypothèse est émise que, alors qu'une rechute ou une réaction immunologiquc dans la lèpre paucibacillairc peut être le résultat de bacilles survivants, celles-ci pourraient être dues à une reinfection dans la lèpre multibacillaire.RESUMEN
En esté estudio con pacientes con lepra aparentemente curada por monoterapia con dapsona, se usó la reacción en cadena de la polimerasa (PCR) para la detección especifia del DNA de Mycoacterium leprae. Se examinaron el esputo y los extendidos de linfa cutánea de 44 pacientes del sanatorio liaba Baghi en Irán. Se usaron iniciadores para el fragmento de 530 pares de bases del gene del antigeno de 36 kD de M. leprae. Los resultados por PCR fueron comparados con los resultados por microscopía para bacilos ácido-resistentes. De las 44 muestras de esputo, dos resultaron positivas por PCR (4.5%) y de los 44 extendidos de linfa cutánea de los mismos pacientes, 10 fueron positivos por PCR (22.7%). Sólo un paciente fue PCR positivo tanto en el esputo como en los extendidos de linfa (2.3%). Por microscopía no se encontraron bacilos ácido-rcsistcntcs. En total, 11 de 44 (25%) pacientes en este estudio resultaron positivos para M. leprae por PCR; se pensó que esto podría indicar la presencia de organismos vivos. Fue particularmente interesante la asociación entre los resultados positivos de los extendidos de linfa cutánea, con la lepra paucibacilar más que con la multibacilar. Se sugiere que mientras que las recaídas o la reacción leprosa en los pacientes paucibacilares pueden resultar de la presencia de bacilos sobrevivientes, en la lepra multibacilar ésta pueden ser el resultado de reinfecciones.Leprosy (Hansen's disease) is a chronic mycobacterial disease, infectious in some cases, which still afflicts millions of people worldwide. Mycobacterium leprae, the etiologic agent of leprosy, is one of the human pathogens which cannot be grown in vitro (6). The long incubation period together with the wide spectrum of clinical manifestations of leprosy have prevented reliable and rapid diagnosis of infections, especially in the tuberculoid and indeterminate forms of the disease (9). Fortunately, new molecular methods have been developed as reliable and sensitive diagnostic tools for the identification of leprosy bacilli. The most significant advance in useful molecular methods, applicable to diagnosis, has been the polymerase chain reaction (PCR).
Multidrug therapy (MDT) was introduced in the Baba Baghi Leprosy Sanatorium of Iran during the 1980s, but some patients living there who had apparently been cured after receiving dapsone monotherapy over many years were not given MDT. It is of interest for many purposes to monitor the bacteriological status of such patients by using the more specific molecular methods now available.
In the study described here, PCR was performed to detect specific M. leprae DNA in specimens from these long-treated leprosy patients by a set of primers with a detection limit of approximately one bacterium (4). The aim of this study was to find out whether PCR detects bacilli that are missed by microscopy, the presence of which might herald relapses in clinical disease.
MATERIALS AND METHODS
Patients and clinical specimens. Fortyfour treated leprosy patients (31 males and 13 females) from the Baba Baghi Leprosy Sanatorium in Azerbaijan, Iran, were selected on the basis of their previous monotherapy with dapsone and entered into the study. Although they seemed to have been cured, they still had physical manifestations of earlier active disease. Recent clinical examinations, however, showed no obvious signs or symptoms of any relapse in their leprosy. Their ages were between 30 and 80 years and, according to their clinical data, over the years 22 were diagnosed with multibacillary (MB) leprosy, 1 with indeterminate leprosy, and 21 with paucibacillary (PB) leprosy. The specimens chosen for study were swabs of skin slits and sputum to provide a basis for comparison with the standard diagnostic method of microscopy. The samples were obtained from the patients as follows:
Slit-skin smears and swabs. Slit-skin smears and swabs were taken from old skin lesions of leprosy. The chosen sites were cleaned with alcohol and allowed to dry. Next, the skin was pinched up into a fold between the index finger and thumb with enough pressure to stop or minimize bleeding. A cut of about 5 mm in length deep enough to penetrate well into the infiltrated layer of the dermis was then made with a small sterile scalpel (6). Smears were prepared on microscope slides and swabs (ENT swabs) were taken from these cuts. After collection, the skin smears were examined in the laboratory. The skin swabs were kept at -20ºC for transport to London for PCR experiments.
Sputum samples. Sputa coughed up by the patients on waking in the morning were collected and smears were prepared on glass slides. These were examined in the laboratory by direct microscopy, and the remaining sputa were transferred to small scrcw-cappcd bottles and kept at - 20ºC for transport to London for PCR examinations.
Acid-fast microscopy. All fixed sputum and slit-skin smears were stained by the Zichl-Ncclsen method for acid-fast bacilli (AFI3) using strong carbol fuchsin, acid-alcohol as a decolorant, and methylene blue as the countcrstain. More than 20 fields of each stained smear were examined carefully under the light microscope using an oil immersion (x 100) lens.
DNA extraction from clinical specimens (1). For skin swab specimens, each was placed in a 10-ml conical tube containing 1 ml phosphate buffered saline (PBS) at pH 7.4 plus a 2x concentration of Fungi-Bact (Gibco) (5), and mixed on a vortcx-mixcr. Next, the specimen was allowed to stand at room temperature overnight, after which it was again vortcx-mixed. In the case of sputum samples, each was liquefied by the method described by Victor, et al. (8). For subsequent DNA extraction, 50 µl of either type of suspension was prepared by the procedure described below.
Diatom suspension (40 µl) was mixed with 900 µl of lysis buffer (containing GuSCN 120 g; 0.1 M Tris-HCl at pH 6.4, 100 ml; 0.2 M EDTA at pH 8.0, 22 ml; and Triton X-100, 2.6 g) in a 1.5-ml Eppendorf microcentrifuge tube, and briefly vortex-mixed. The clinical specimen (50 id) was added to the above, vortex-mixed for 5 sec, and allowed to stand at room temperature for 10 min. Next, it was vortex-mixed again, and spun at 12,000 x g x 15 sec. The supernatant was discarded, and the nucleic acid (NA)-pellet was washed twice with washing buffer (containing GuSCN 120 g, and 0.1 M Tris-HCl at pH 6.4, 100 ml), twice with 70% ethanol, and once with acetone. The acetone was removed and the NA-pellet was dried at 56ºC for 10 min; 100 ,d of TE buffer (Tris-HCl at pH 8.0, 10 raM and EDTA at pH 8.0, 1 mM) were added to the NA-pellet, vortex-mixed, and incubated for 10 min at 56ºC. It was mixed again, spun at 12,000 x g x 2 min, and 5 μl of the supernatant was used for PCR.
Preparation of chromosomal DNA. M. leprae DNA was isolated from dead M. leprae (1.25 x 109 AFB/ml, killed by radiation) by the method mentioned previously (1), and was used as a positive control in each set of PCR amplifications.
Selection of primers. The primers used for amplification were designed by Hartskeerl, et al. (4) and were selected on the basis of the nucleotide sequence of the gene encoding the 36-kDa antigen of M. leprae. The primers were S13 and S62. The sequence of the primers (synthesized by Oswell DNA Service, Edinburgh, U.K.), which amplify a 530-bp fragment of the M. leprae DNA sequence, were S13 (5'-CTCCACCT-GGACCGGCGAT-3') and S62 (5'-GAC-TAGCCTGCCAAGTCG-3').
PCR procedure. Briefly, 5 µl of each DNA extract was incubated in a 45-µl reaction mixture containing 100 mM Tris-HCl (pH 8.3), 500 mM KC1, 15 mM MgCl2, 0.1% gelatin, 1 each of primers SI3 and S62, 0.2 mM each of deoxynuclcotides dATP, dCTP, dGTP and dTTP (Pharmacia), and 2.5 units of Taq polymerase (purchased from Perkin-ElmerCctus). The reaction mixtures were covered with 40 µl of sterile mineral oil. A control tube containing no target DNA as a negative control and another tube containing chromosomal DNA of M. leprae as a positive control were included with every set of tests. Precautions were taken to avoid contamination with extraneous DNA. In order to test for the presence of PCR inhibitors, PCR-negative specimens were re-tested after adding 2 ,ul of the chromosomal DNA to the amplification mixture (2).
The reaction was performed using an automated thermal cycler and 45 amplification cycles were performed. Each cycle consisted of denaturation at 94ºC for 2 min, annealing of primers at 55ºC for 2 min, and primer extension at 72ºC for 3 min. After the 45th cycle, the extension reaction was continued for another 12 min at 72ºC (4). The presence of the 530-bp amplification product was sought by electrophoresis of 5 µl of the amplified mixture at 6V/cm for 40 min on an agarose gel (0.8%). The DNA was stained with ethidium bromide and visualized on a 302-nm ultraviolet transilluminator (3). The molecular size marker used was 0 X 174 DNA Hae III digest (Sigma).
RESULTS
A total of 88 clinical specimens from 44 patients were examined under the microscope after staining for AFB and none were seen. In contrast, PCR detected the presence of the 530-bp DNA fragment specific for leprosy bacilli in 12 (13.6%) of the specimens. No PCR inhibitors were found in any of the PCR-ncgativc samples. Ten of the 44 slit-skin swabs were positive and 2 of the 44 sputum samples were positive by PCR. In one patient PCR was positive for both skin and sputum, thus material from M. leprae was detected in a total of 11/44 patients. Among male patients, 7/31 were PCR positive (including the patient positive in both samples); among females, 4/13 were positive.
The distribution according to type of the original disease is shown in Table 1. The positivity rate of PCR for PB patients (38.1%) was about three times higher than for MB patients (13.6%). The distribution according to age and leprosy type and the PCR results are shown in Table 2. The greatest positivity rate was found in the youngest age group, those aged between 30 and 45 years, although the group was small.
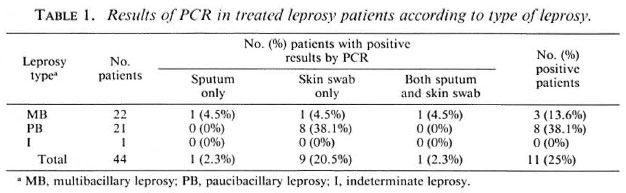
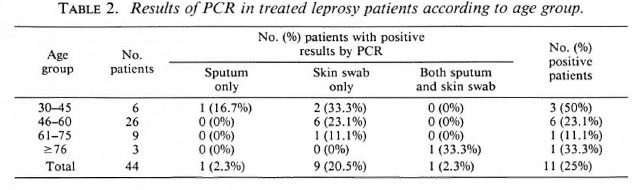
In total, 25% of the treated leprosy patients in this study were found to be PCR positive for M. leprae by this technique.
DISCUSSION
The primers SI3 and S62 were chosen on the basis of their selecting a nucleotide sequence of a gene encoding the 36-kDa antigen of M. leprae. A 530-bp fragment of the sequence that is specific for M. leprae can be amplified. The significant specificity of this set of primers means that they give no amplification of human DNA or of DNA from a number of other bacteria which may be present in human-derived samples (4). Thus, this method is highly useful for the accurate detection of leprosy bacilli in human specimens. The detection limit of this PCR with primers S13 and S62 is reported to be 1 to 10 bacilli (4), and recently M. leprae DNA from an ancient bone dating from 600 A.D. has been identified by their use (7). Theoretically this technique should be much more sensitive than other methods, such as microscopy for the direct detection of M. leprae.
Our finding of 11/44 previously treated leprosy patients still positive by PCR for M. leprae-spccific DNA is surprising since all the patients in this study were thought to have been cured by dapsone monotherapy before 1982. Although it is possible that the bacilli from which the DNA was amplified were dead, and our own data (7) on its survival in ancient bones raises this possibility, it would be surprising if such long pieces of DNA could survive in living host tissues for long after the bacilli were killed and even their acid-fast ghosts had disappeared. The greater PCR positivity of patients with PB disease over those with MB leprosy also makes it unlikely that the DNA came from dead bacilli, since numbers of dead bacilli might be expected to be much greater in the MB patients.
Detection of DNA of M. leprae in slitskin swabs really does suggest that live bacilli are still present, perhaps as pcrsistcrs, held in a state of virtually suspended animation by the immune mechanisms of the host. A study reported by the World Health Organization (WHO) (10) showed that in a small proportion of patients properly treated with dapsonc monotherapy pcrsistcrs may survive for as long as 20 years. They might also be vegetative organisms about to lead to relapse. It is more difficult to be sure of the significance of positive results on sputum samples, since these could more readily be from external contamination.
The surprise finding, of more positive results on slit-skin swabs from patients who had PB disease than from those who had MB disease, reaches statistical significance (p < 0.03). An obvious explanation for this is the likely difference in treatment given to those with the two types of leprosy.
Whereas MB leprosy was treated for several years with high-dose dapsonc, PB patients often were given shorter treatment with low-dose dapsonc. Once a PB patient has apparently overcome the infection, chemotherapy usually can be stopped with impunity, since active disease rarely recurs. Late reactions of the reversal type occurring in a proportion of such patients are explained as due to residual immunological responses to remaining antigen. Our results suggest a different explanation. A correlation might be sought between the occurrence of late reactions in PB patients and the detection of M. leprae DNA in their tissues.
In reality, the result of a short course of dapsonc for PB disease may be to slow the metabolic activity of the bacilli and perhaps to kill a proportion of them, allowing the immune system to regain control of the situation, returing the infection to a latent form in which the bacilli continue as pcrsistcrs in a proportion of patients. This would not occur in the treatment of MB disease, where the patient seems to lack the immunological power to hold the organisms in a persister state, and all have to be killed by the chemotherapy to stop reactivation from occurring. Such patients, after cessation of chemotherapy, may return to a state of high susceptibility to re-infection. The 2/22 MB patients found to be PCR positive in this study may be re-infected patients progressing toward a relapse of their disease.
Table 2 shows that younger patients are more likely to have detectable M. leprae DNA in their tissues than are older patients. This may be due to age but might also be due to the preponderance of PB disease in the younger group, which the data suggest.
The results illustrate the potential value of PCR in the investigation of leprosy, and particularly indicate that it may be a useful tool for confirming that the bacterial load actually has been removed by treatment in PB disease. We are hoping to extend our studies to patients receiving modern multidrug therapy to discover whether the proportion of PB patients remaining PCR positive after treatment includes those who suffer damaging late reactions.
Acknowledgment. We sincerely thank Dr. M. H. Mobayen, the Director, and Dr. F. Fcval, the leprologist. Baba Baghi Leprosy Hospital and Sanatorium, Iran, and Mr. Ali Akhavan, Assistant, Research Department, Tabriz University of Medical Sciences, for their valuable help throughout this study. Dr. P. R. Klatser's advice on swab sampling of skin slits is gratefully acknowledged, as is his supply of suitable swabs.
REFERENCES
1. BOOM, R., SOL, C. J. A., SALIMANS, M. M. M., JANSEN, C. L., WERTHEIM-VAN DILLEN, P. M. E. and VAN DER NOORDAA, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28(1990)495-503.
2. DE WIT, M. Y. L., FABER, W. R., KRIEG, S. R., DOUGLAS, J. T., LUCAS, S. B., ASUWAT, N. M., PATTYN, S. R., HUSSEIN, R., PONNIGHAUS, J. M., HARTSKEERL, R. A. and KLATSER, P. R. Application of a polymerase chain reaction for the detection of Mycobacterium leprae in skin tissues. J. Clin. Microbiol. 29(1991)906-910.
3. EISENACH, K. D., CAVE, M. D., BATES, J. H. and CRAWFORD, J. T. Polymerase chain reaction amplification of repetitive DNA sequence specific for Mycobacterium tuberculosis. J. Infect. Dis. 161(1990)977-981.
4. HARTSKEERL, R. A., DE WIT, M. Y. L. and KLATSER, P. R. Polymerase chain reaction for the detection of Mycobacterium leprae. J. Gen. Microbiol. 135(1989)2357-2364.
5. INNIS, M. A., GELFAND, D. H., SNINSKY, J. J. and WHITE, T. J. PCR Protocols: a Guide to Methods and Applications. San Diego, California: Academic Press, 1990, pp. 147-148.
6. PARKER, M. T. and COLLIER, L. H. Topley& Wilson 's Principles of Bacteriology. Virology and Immunity. Volume3. 8th cdu. Scvcnoaks, Kent, U.K.: Edward Arnold, 1990, pp. 82-84.
7. RAH, A., SPIGELMAN, M., STANFORD, J., LEMMA, E., DONOGHUE, H. and ZIAS, J. Mycobacterium leprae DNA from ancient bone detected by PCR. Lancet 343(1994)1360-1361.
8. VICTOR, T., DU TOIT, R. and VAN HELDEN, P. D. Purification of sputum samples through sucrose improves detection of Mycobacterium tuberculosis by polymerase chain reaction. J. Clin. Microbiol. 30(1992 )1514-1517.
9. WHO STUDY GROUP. Chemotherapy of leprosy for control programmes. Geneva: World Health Organization, 1982 . Tech. Rep. Ser. 675.
10. WHO EXPERT COMMITTEE ON LEPROSY. Sixth report. Geneva: World Health Organization, 1988 . Tech. Rep. Ser. 768.
1. M.Sc.; University College London Medical School, Department of Medical Microbiology, Division of Bacteriology, 67-73 Riding House Street, London W1P 7PN, U.K.
2. Ph.D.; University College London Medical School, Department of Medical Microbiology, Division of Bacteriology, 67-73 Riding House Street, London W1P 7PN, U.K.
3. Ph.D., University College London Medical School, Department of Medical Microbiology, Division of Bacteriology, 67-73 Riding House Street, London W1P 7PN, U.K.
Reprint requests to Dr. Stanford.
Received for publication on 15 July 1994.
Accepted for publication on 30 August 1994.