- Volume 62 , Number 4
- Page: 499–511
Genetic control of susceptibility to leprosy in French Polynesia; no evidence for linkage with markers on telomeric human chromosome 2
ABSTRACT
Several lines of evidence have suggested a role of genetic factors in susceptibility to leprosy. In the mouse, natural susceptibility to infection with mycobacteria is controlled by the chromosome 1 Bcg locus, a region which is syntenic with a fragment of the human chromosome 2q, region q31-q37. It has been postulated that a human homolog of the Beg gene controls susceptibility to leprosy per se, and may be located on chromosome 2q. In order to test the influence of this putative gene on leprosy per se, we performed linkage analyses in a set of seven multicase French Polynesian pedigrees, using an affected sib pair method and the LOD score method employing different modes of inheritance. Family members were typed for eight polymorphic loci on chromosome 2q:CRYGP1, FN, TNP1, VIL, DES, INH, PAX3, and UGT1A1. Our data provide evidence against the presence of a gene controlling susceptibility to leprosy per se on human chromosome 2q in the French Polynesian population.
RÉSUMÉ
Divers indices ont suggéré un role de facteurs génétiques dans la susceptibilité à la lèpre. Chez la souris, la susceptibilité naturelle à l'infection par des mycobactéries est sous le controle du locus Bcg du chromosome 1, une région synténique avec un fragment du chromosome humain 2q, région q31-q37. On a émis l'hypothèse qu'un homologue humain du gène Bcg contrôle la susceptibilité à la lèpre per se, et pourrait être localisé sur le chromosome 2q. Afin de tester l'influence de ce gène supposé sur la lèpre per se, nous avons réalisé des analyses de lien dans un ensemble de sept généalogies à cas multiples de Polynésie française, sur base d'une méthode des paires jumelles et de la méthode du score LOD employant différents modes d'héritage. Les membres des familles ont été typés en fonction de huit loci polymorphiques sur le chromosome 2q: CRYGP1, FN, TNP1, VIL, DES, INH, PAX3 et UGT1A1. Nos données plaident contre la présence d'un gène contrôlant la susceptibilité à la lèpre per se sur le chromosome humain 2q dans la population de Polynésie française.RESUMEN
Hay varias lincas de evidencia que sugieren la participación de factores genéticos en la susceptibilidad a la lepra. En el ratón, la susceptibilidad natural a la infección por micobacterias está controlada por el locus Bcg del cromosoma 1, una región análoga a la región q31-q37 del cromosoma 2q humano. Se ha propuesto que un gene análogo al gene Bcg controla la susceptibilidad a la lepra per se. y que este gene podría estar localizado en el cromosoma 2q. Para probar la influencia de este gene putativo sobre la lepra per se, se hizo un análisis de ligamiento en 7 familias de la Polinesia francesa con casos múltiples de lepra, utilizando los métodos de análisis genético apropiados. Los miembros familiares se tipificaron en función de 8 loci polimórficos del cromosoma 2q: CRYGP1, FN, TNP1, VIL, DES, INH, PAX3, y UGT1A1. Los resultados no dieron evidencias de la presencia de genes, en el cromosoma 2q, que controlen la susceptibilidad a la lepra en la población de la Polinesia francesa.Leprosy is a chronic infectious disease caused by Mycobacterium leprae and afflicts an estimated 5.5 million people in the world (24). This disease can cause permanent and extensive deformities of the skin and peripheral nerves and, thus, has serious social and economic consequences. Most individuals infected with M. leprae develop effective immunity without clinically evident forms of the disease; whereas those people who develop leprosy present different spectral manifestations, ranging from the tuberculoid to the lepromatous pole accompanied by a gradual decrease of cell mediated immunity (34).
The possible role of the host genetic background in resistance and susceptibility to leprosy is supported by several lines of epidemiological evidence. These include the variable incidence rate of leprosy among certain ethnic groups (3,43) and the restriction of the disease to certain familial clusters living in endemic areas (3,31,40). In addition, the concordance rate for leprosy was shown to be significantly higher among monozygotic twins (60%-85%) than among dizygotic twins (15%-20%) (6,23). Segregation analyses have provided more conclusive evidence regarding the influence of genetic factors and the mode of inheritance of the disease trait in the etiology of leprosy (1,17,42,48). The mode of inheritance has been found to vary among populations. For all forms of leprosy (leprosy per se) a nearly dominant mode of inheritance was found in Thailand (48), while a recessive or codominant mode of inheritance was found in Desirade Island (1). In The Philippines, segregation of a recessive susceptibility gene was found for lepromatous leprosy (42), while recessive inheritance of nonlepromatous leprosy was concluded in India (17). The complex segregation analysis performed in the Desirade study also revealed the presence of incomplete penetrance and phenocopies. Overall, predisposition to leprosy appears to be of a multifactorial nature with at least two groups of genes participating in the control of the disease: non-HLA (human leukocyte antigen)-linked genes which determine susceptibility to leprosy per se and HLA-linked genes which are implicated in the spectral clinical manifestations of the disease (1). Population and family studies have shown that tuberculoid leprosy was associated with HLA-DR2 in India (8), in Japan (22) and in Thailand (35) or with HLA-DR3 in Surinam (28), while lepromatous leprosy was associated with HLA-DQw1 in Venezuela (29,44). In the mouse, innate resistance and susceptibility to several pathogens including mycobacteria is controlled by a single autosomal gene termed Beg which is expressed in two allelic forms, the dominant resistance allele and the recessive susceptibility allele (16). A high resolution genetic map of the Beg region on proximal mouse chromosome 1 has revealed the gene order: centromerc- Col3a1-Cryg-Fn-1-Tp-1-Vil/Bcg-Des-Inha-Pax3-Akp3-Acrg-Sag-Col6a3 (11,20,21,36-38). A35-cM fragment around the murine Beg locus (from Col3a1 to Col6a3) has been conserved between the murine chromosome 1 and the telomeric end of human chromosome 2q, region 2q32-2q37 (38). Moreover, the gene order appears to be conserved among this group of syntenic loci (38,39). Thus, a human homolog for the Beg locus could be located on chromosome 2q, region q32-37, which defines the main target area for the search of human Bcg homolog.
In French Polynesia, a monitoring and prevention policy for leprosy control has been implemented since the end of the 1940s. The prevalence rate decreased from 2.40/1000 in 1946 to 0.48/1000 in 1987. Despite this decline, leprosy is still endemic and considered as a public health problem (5). Furthermore, French Polynesia represents a geographic area where the history of leprosy families is well documented.
The objective of this study was to investigate, in French Polynesian families, the presence of a gene that could determine susceptibility to leprosy and be located on human chromosome 2q using a sib-pair method and the logarithm of odds (LOD) score method.
MATERIALS AND METHODS
Patients and families
Seven multicase pedigrees with two (4 families) or three (3 families) generations and with at least one affected parent were identified in 1990. Families were identified from the records of the Institut Territorial Louis Malardé in Tahiti, French Polynesia, where a central register has kept all information on leprosy patients since 1902. For each leprosy patient, a file is kept indicating civil status, date and place of leprosy detection as well as the results of clinical, microbiological and pathological examination and the nature and duration of treatment. Leprosy patients are classified according to the Ridley-Jopling classification (32). Since 1982, multidrug therapy (MDT) has been implemented with daily administration of dapsone and rifampin for paucibacillary patients and a daily supplement of clofazimine during the first 12 months and of ethionamide for the first 2 months for multibacillary cases of leprosy.
All families were examined for possible nonpaternity by showing the segregation of the blood groups, the highly polymorphic genetic markers D2S53 (25) and D2S104 (10,49), and other markers used in this study. Inconsistencies were found for four individuals, who were excluded from the study.
The entire study group of 84 individuals comprised a total of 39 leprosy patients, including 19 paucibacillary (indeterminate, IBT, TT forms) and 20 multibacillary (BL and LL forms) patients. The leprosy patients comprised 24 males and 15 females with a mean age at onset of 27 years.
DNA preparation
A 10-ml, heparinized peripheral blood sample was collected from each patient following informed consent. Lymphocytes were immortalized by transformation with Epstein-Barr virus (2). Genomic DNA was prepared according to standard protocols using SDS lysis, proteinase K digestion, phenol/ chloroform extraction and ethanol precipitation (33).
Typing
Individual family members were typed at 8 polymorphic loci (18 allelic systems) located on human chromosome 2q (Table 1). These marker loci were previously mapped on mouse chromosome 1 (11,20,36-38,45).
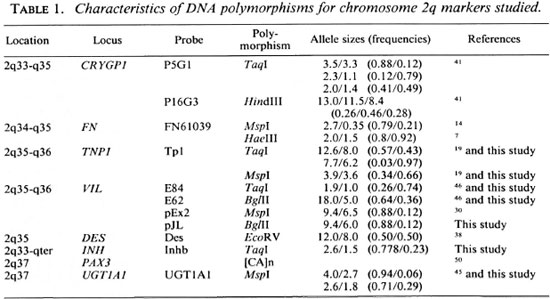
Restriction fragment length polymorphism (RFLP) analysis. Five µ g samples of DNA were digested to completion with 50 U of the appropriate restriction enzyme according to the recommendations of the manufacturer (Pharmacia; New England Biolabs, Beverly, Massachusetts, U.S.A.). The DNA fragments were separated in a 1% agarose gel, except for Hae III digested fragments which were separated in a 1.4% agarose gel. Gels were transferred to N-Hybond membranes (Amersham, U.K.) as previously described (33). Southern blots were prehybridized at 65ºC in hybridization solution (6x SSC, 5x Denardt's, 0.1% SDS and 100 mg/ml boiled salmon sperm DNA).
For mouse exon probes E84 or E62, prehybridization and hybridization were at 54ºC using the hybridization solution described above. The probes described in Table 1 were labelled to a high specific activity (5 x 108 cpm/ µ g DNA) with [ α -32P-dATP] (Dupont, NEN Research Products, Boston, Massachusetts, U.S.A.) using the random hexanucleotide method (12). Membranes were washed as follows: twice with 2 x SSC and 0.1% SDS at room temperature for 15 min and twice with 0.5 x SSC and 0.1% SDS at the hybridization temperature for 15 min.
Membranes were exposed for 2 days at -70ºC, using Kodak XAR films and Dupont Lightning-Plus intensifying screens.
Polymorphism analysis of PAX3. [CA] dinucleotide repeat polymorphisms were detected for the PAX3 gene after PCR amplification (50). Amplification was performed in a total volume of 15 µ containing 250 ng of genomic DNA, 9 pmol of each primer, 0.05 mM TrisHCl pH8, 1 mM MgC12, 0.05% Nonidet P40, 0.005% Tween 20, 200 µ M each of dCTP, dGTP, and dTTP, 30 µ M dATP, 0.6 µ Ci 35S dATP and 3.5 units of Taq polymerase. Samples were processed through one step of denaturation (94ºC for 5 min), followed by 40 cycles of denaturation (94ºC for 1 min), annealing (54ºC for 1 min), elongation (72ºC for 1 min) and a last step of elongation (72ºC for 7 min). Amplification products were electrophoresed for 3-4 hr on a 6% polyacrylamide denaturing gel. Gels were dried and exposed for 2 days at room temperature on Kodak XAR-film.
All genotypes were interpreted from the autoradiographs by two independent observers.
Linkage analyses
Linkage analyses were carried out using a sib-pair method and the LOD score method. For the analyses, susceptibility to leprosy per se (any form of leprosy) was used as the phenotype to be analyzed.
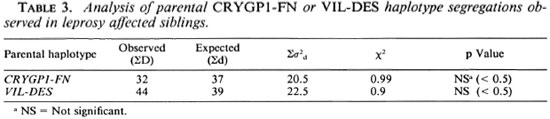
Sib-pair method. The nonparamctric statistical method described by de Vries, et al. (9) was used to detect the segregation of parental haplotypes in sibships with at least two afTected siblings. The observed difference (D) between the number of afTected siblings with one and those with the other parental haplotype was compared with the expected difference (d) for random segregation using the statistic test: (|ΣD - Σd| - ½ )2/Σσd2 (σd2 is the variance of d and Σ d and Σ D the summations of d and D values for all sibships). This test has a chi-squared distribution of 1 df. Two different haplotypes were tested: the haplotype including CRYGP1 and FN and the haplotype including VIL and DES. These haplotypes were chosen because of the absence of recombination events, except for one patient who was excluded from the analysis with the CR YGP1-FN haplotype and another for the analysis with the VIL-DES haplotype. For these analyses, the pedigrees were divided into nuclear families. Nuclear families with at least one heterozygous parent were tested: the CRYGP1-FN haplotype was assessed in 7 and the VIL-DES haplotype in 8 multiple-case nuclear families. All possible sib pairs were considered when more than two affected siblings were present.
LOD score method. Linkage analysis was carried out using the LINKAGE 5.1 computer program package (18). Two point LOD scores were calculated with the MLINK program using marker haplotypes for each locus, except for the CRYGP1 locus. For this locus, recombination events have been detected between CRYGP1 (P5G1) and CRYGP1 (P16G3) (E. Schurr, unpublished data). LOD scores obtained at different recombination fractions (Θ = 0 to Θ = 0.5) were added for all families. A LOD score of 3.0 is considered as significant evidence in favor of linkage, and a LOD score of - 2.0 as evidence against linkage between the markers and the putative disease susceptibility locus at a given Θ value. From the Central Register, wc determined the number of leprosy cases (afTected or having been affected) in the French Polynesian population (2.22/1000). Under the assumption of Hardy-Weinbcrg equilibrium, we defined the disease gene frequency for each mode of inheritance: 0.0471 for the recessive, 0.0022 for the codominant, and 0.0011 for the dominant mode of inheritance.
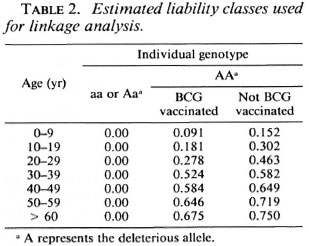
Successive linkage analyses were performed: a) The three modes of inheritance with a range of assumed penetrance values from 100% to 40% for the susceptible allele as well as with affecteds only were compared, b) In addition, two-point analysis of the data was carried out using a complex model. The hypothesis was that a human homolog of the Bcg gene was located on chromosome 2q and that recessive alleles of this locus conferred susceptibility to leprosy with incomplete penetrance as suggested by Abel and Demenais (1). For heterozygotes, we assumed that the susceptibility allele was not expressed. The penetrance was considered to vary with age and BCG vaccination. Since leprosy has a variable age at onset, 10-year liability classes (frequencies of infection) were defined from the general population and the data recorded in the Central Register. Individuals were grouped by attained age in 1990 or at death. Different studies have shown a variation in the protective efficacity of BCG vaccination against leprosy ranging from 20% to 80% (13). A study carried out in New Guinea indicated a protection rate of about 40% for people under 30 years old (4). Since the New Guinea population and the French Polynesian population could be racially related, we assumed a 40% BCG protection rate for people under 30 years old and 10% for those older. Individuals were examined for evidence of BCG vaccination. For deceased people, the presence or absence of BCG vaccination was determined by medical records or interviewing the family members. Table 2 presents the parameters used for this analysis. c) By considering all leprosy cases without affected relatives as nongenetic causes, we defined a 0.48 phenocopy rate. This parameter was introduced in the complex model described in b above. Allele frequencies for the RFLP markers listed in Table 1 were determined in 38 chromosomes from unrelated Polynesian individuals. For the PAX3 marker, we considered 10 alleles with equal frequencies. Recombination frequencies were assumed to be equal in males and females.
RESULTS
In the mouse, the Beg gene controls innate resistance to mycobacterial infections and is located on a syntenic linkage group which is conserved on human chromosome 2q (38). Therefore, we assumed that the human homolog of the Beg gene was located on human chromosome 2q, and we evaluated 8 loci (18 allelic systems) for possible linkage with susceptibility to leprosy per se. The DNA of 84 patients from 7 multicase families (The Figure) was typed for polymorphic chromosome 2 markers using [CA]n repeats and RFLP analysis. The allele frequencies for the marker loci are presented in Table 1. Since the gene order on the syntenic fragment appears conserved between mouse and man (38), we constructed haplotypes using the following order: CRYGP1-FN-TNP1- VIL-DES- INH-PAX3-UGT1A1. For deceased individuals, haplotypes were deduced from the haplotypes of their offsprings and/or their parents.
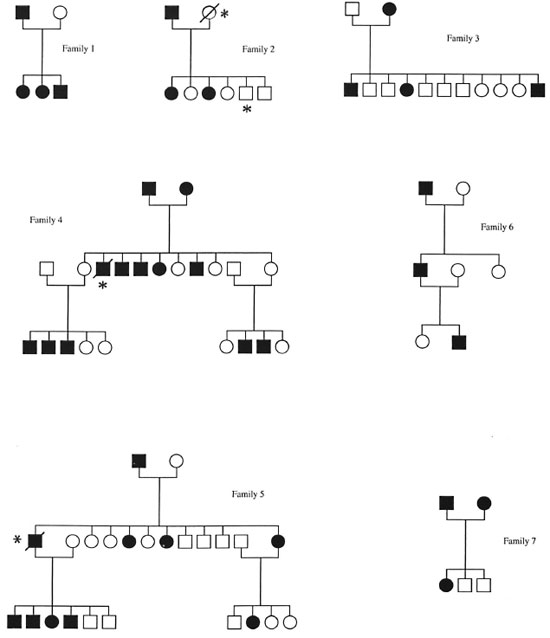
The Figure. Pedigrees of the seven families analyzed in this study. Solid symbol = affected subject; slash ; subject deceased; * = subject not typed.
Results with nonparametric method
The affected sib-pair method used in this study is based on the comparison of expected and observed identical by descent distribution. Two pairs and 11 trios were available for the CRYGP1-FN haplotype and 3 pairs and 7 trios for the VIL-DES haplotype. The probability of random parental haplotype segregation was calculated in Table 3 according to the statistical test described by dc Vries, et al. (9). No significant deviation from random segregation was observed for the two haplotypes tested (p < 0.5).
Results with LOD score method
Although no evidence for the presence of a gene located on chromosome 2q conferring susceptibility to leprosy per se was suggested with the sib-pair method, we carried out LOD score analyses in order to confirm this result. We first tested the three genetic models assuming a penetrance of 100%, 80% and 40% for the susceptible alleles. In addition, we used a penetrance-free model with affecteds only. Tables 4, 5 and 6 compare the LOD score values obtained for linkage between the putative leprosy susceptibility gene and all the gene markers. The LOD scores are predominantly negative regardless of the mode of inheritance and penetrance values. Under the recessive model, whatever the penetrance values assumed (100%, 80% and 40%), data provide evidence against the existence of a putative gene falling within 10 cM to 20 cM ofVIL(LOD score value of -2 obtained at a recombination fraction between 0.1 -0.2). Under the dominant model, there is also evidence against a putative gene falling within 5 cM to 20 cM of TNP1 (LOD score value of -2 obtained at a recombination fraction of 0.05-0.2). Considering the codominant model, data provide evidence against a putative gene falling within 5 cM to 10 cM of TNP1 with penetrance values of 100% and 80% (LOD score value of -2 obtained at a recombination fraction of 0.05-0.1). Results obtained with affecteds only are not conclusive assuming a recessive mode of inheritance of susceptibility to leprosy per se (LOD score values between -2 and +3).
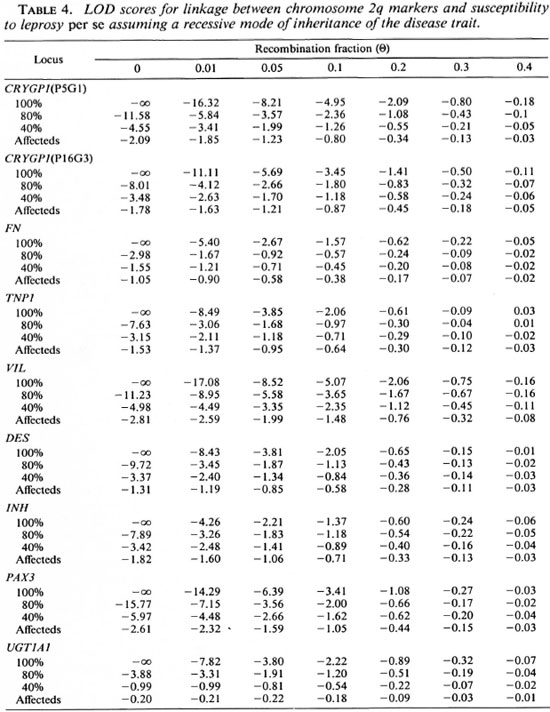
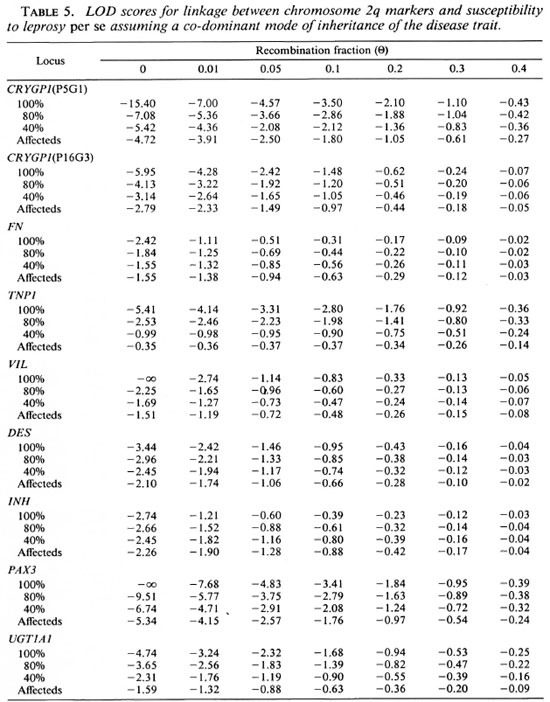
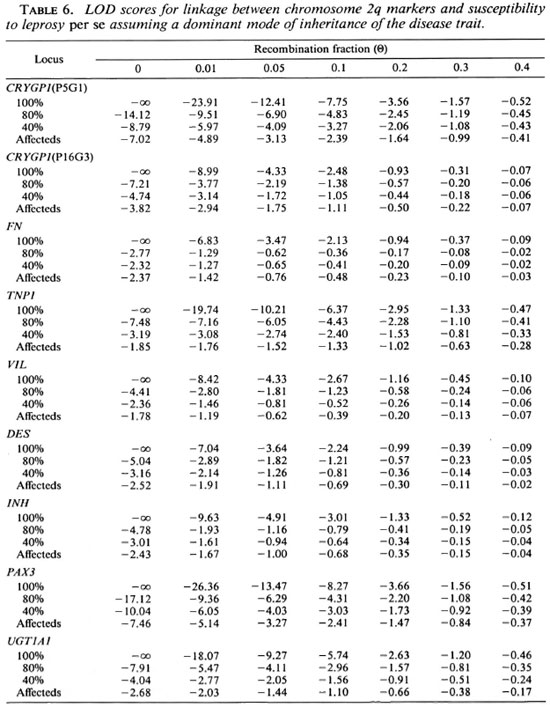
Under a co-dominant or dominant model, there is no convincing evidence for the presence of a putative disease gene falling within 5 cM to 20 cM of the distal markers CRYGP1 and PAX3. The reduction of the penetrance for the susceptible allele or analyses with affecteds only reduce the LOD score values, but these values remain predominantly negative and provide consistent evidence against linkage.
Although the results with simple models provide evidence against the presence of a gene controlling the susceptibility to leprosy assumptions are presented in Table 6. Again, per se on human chromosome 2q, we tested the LOD score values are predominantly a complex model. The assumptions for this negative. Evidence against the co-segregation of VIL with a gene controlling the suceptibility to leprosy per se within this region was provided (LOD score of -2 obtained at a recombination fraction between 0.1 and 0.2). The introduction of a phenocopy rate of 0.48 did not significantly modify these results (data not shown).
DISCUSSION
To test whether an innate susceptibility locus controlling susceptibility to leprosy is located on human chromosome 2q, the putative site of the human Bcg homolog, we have performed linkage analyses in multiple affected families of French Polynesia, a leprosy-endemic region. The results of our study do not provide evidence for the association between leprosy per se and markers located on telomeric human chromosome 2q in these Tamilies.
Leprosy is a complex trait and, thus, linkage analysis of the disease is complicated by many factors including reduced penetrance, genetic heterogeneity of the trait, gene interactions, and presence of phenocopies (26). To circumvent some of these difficulties, we tested a nonparametric sib-pair method, simple LOD score analysis models under different inheritance modes, and carried out linkage analyses using affecteds only. The nonparametric affected sib-pair methods do not require specification of the genetic model if both parents are available for analysis (27), and they are robust to detect disease susceptibility loci which may have major or minor effects. However, the sample in our study was not of sufficient size to detect modifier effects of chromosome 2q loci. Our results with the affected sib-pair method described by de Vries, et al. (9), uses identity by descent and did not demonstrate linkage between CRYGP1, FN, VIL and DES and the disease phenotype. A second strategy we employed to detect linkage, provided the disease gene has a strong enough effect to be detected despite a possibly erroneous assumption made in the analysis, was to carry out LOD score analysis under simple modes of inheritance. We have tested three modes of inheritance with full and reduced penetrance. However, evidence remains consistently against the presence of a gene controlling leprosy per se on the region studied. Another strategy was to perform LOD score analysis using affecteds only to avoid the problem of reduced penetrance. Our results using this approach still did not provide any influence of a putative disease susceptibility locus in the region 2q33-q37. Finally, a complex model of linkage analysis was performed. In this model, we assumed a recessive mode of inheritance of susceptibility to leprosy per se as well as an age-and BCG-vaccination-dependent penetrance. Again, the results provide evidence against the presence in this region of a gene controlling susceptibility to leprosy per se.
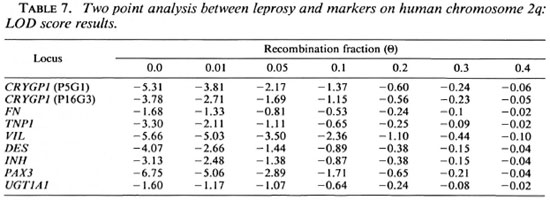
A previous linkage analysis has been performed with 17 multicase Pakistani and Brazilian families typed for 9 markers in the 2q33-q37 region, including FN, TNP1, VIL and DES (39). The results of this study also excluded the presence of a putative disease susceptibility gene on the 2q33-q37 region, assuming a recessive mode of inheritance of susceptibility to leprosy per se and 100% penetrance. With reduced penetrance (80% and 60%) for the susceptibility allele, the presence of such a gene was excluded within the TNP1-DES region. Our data are in agreement with these observations.
Although our results did not reveal the influence of a putative gene in the chromosome 2q region on susceptibility to leprosy per se in the French Polynesian population, leprosy is a complex trait which could have a genetically different etiology in different families. With the low number of families included in this study, no genetic heterogeneity could be detected. Thus, it is possible that genetic linkage could have been masked by undetected heterogeneity (27). If only a proportion of the families (10% to 30%) arc linked to a marker, the families that are unlinked will give negative evidence against linkage (15). One way to circumvent heterogeneity is to use large, multigenerational pedigrees so that only one form of the disease is likely to segregate.
Recently, a candidate gene for the murine Bcg gene, the Nramp gene, has been cloned (46). This gene encodes a putative integral membrane protein. Susceptibility to infection with M. bovis (BCG) seems to be associated with a substitution in the predicted second transmembrane domain. Additional linkage analyses between leprosy and the human homolog of this candidate gene in the French Polynesian population should confirm or refute the results of our linkage analysis.
Acknowledgment. The authors wish to thank all of the patients and their families who have participated in this study. They also thank Jeannette Mahaa, Dr. Philippe Glaziou and Dr. Jean-Louis Cartel for their help and contributions to this work. This work was supported by a grant from Association Raoul Follereau.
REFERENCES
1. ABEL, L. and DÉMENAIS, F. Detection of major genes for susceptibility to leprosy and its subtypes in a Caribbean island: Dcsiradc Island. J. Hum. Genet. 42(1988)256-266.
2. ANDERSON, M. A. and GUSELLA, J. F. Use of cyclosporin A in establishing Epstein-Barr virus transformed human lymphoblastoid cell lines. In Vitro 20(1984)856-868.
3. AYCOCK , W. L. Familial susceptibility as a factor in the propagation of leprosy in North America. Int. J. Lepr. 8(1940)137-150.
4. BAGSHAWE, A., SCOTT, G. C, RUSSEL, D. A., WIGLEY, S. C, MERIANOS, A. and BERRY, G. BCG vaccination in leprosy: final result of the trial in Karamui, Papua New Guinea. Bull. WHO 67(1989) 389-399.
5. CARTEL, J.-L., BOUTIN, J. P., SPIEGEL, A., GLAZIOU, P., PUCHART, R., CARDINES, R. and GROSSET, J.-H. Leprosy in French Polynesia; epidemiological trends between 1946 and 1987. Lepr. Rev. 63(1992)211-222.
6. CHAKRAVARTTI, M. R. and VOGEL, F. A twin study on leprosy. Top. Hum. Genet. 1(1973)1-143.
7. COLOMB, I. M., GARDELLA, R., BARLATI, S. and VAHERI, A. A frequent Haelll RFLP of the human fibronectin gene. Nucleic Acids Res. 16(1987)6761.
8. DE VRIES, R. R., MEHRA, N. K., VAIDYA, M. C, GUPTE, M. D., MEERA KHAN, P. and VAN ROOD, J. J. HLA-linked control of susceptibility to tuberculoid leprosy and association with HLA-DR types. Tissue Antigens 16(1980)294-304.
9. DE VRIES, R. R. P., NUENHUIS, L. E., LAI A FAT, R. F. M. and VA N ROOD, J. J. HLA-linked genetic control of host response to Mycobacterium leprae. Lancet 2(1976)1328-1332.
10. DONNIS-KELLER, H. NIH/CEPH collaborative mapping group; a comprehensive genetic linkage map of human genome. Science 258(1992)148-152.
11. EPSTEIN, D. J., VEKEMANS, M. and MALO, D. splotch (Sp211), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell 67(1991)767-774.
12. FEINBERG, A. P. and VOGELSTEIN, 13. High specific activity labeling of DNA restriction endonuclease fragments: addendum. Anal. Biochem. 137(1984)266-267.
13. FINE, P. E. M. BCG vaccination against tuberculosis and leprosy. Br. Med. Bull. 44(1988)691-703.
14. GARDELLA, R., COLOMBI, M. and BARLATI, S. A common Msp\ RFLP of the human fibronectin gene (FN1). Nucleic Acids Res. 16(1988)1651.
15. GREENBERG, D. A. There is more than one way to collect data for linkage analysis. What a study of epilepsy can tell us about linkage strategy for psychiatric disease. Arch. Gen. Psychiatry 49(1992)745-750.
16. GROS, P., SKAMENE, E. and FORGET, A. Genetic control of natural resistance to Mycobacterium bovis (BCG) in mice. J. Immunol. 127(1981)2417-2421.
17. HAILE, R. W., ISELIUS, L., FINE, P. E. and MONTON, N. E. Segregation and analysis of 72 leprosy pedigrees. Hum. Hered. 35(1985)43-52.
18. LATHROP, G. M., LALOUEL, J. M., JULIER, C. and OTT, J. Strategics for multilocus linkage analysis in humans. Proc. Natl. Acad. Sci. U.S.A. 81(1984)3443-3446.
19. LUERSSEN, H., MATTEI, M. G., SCHOTER, M., GRZESCHIK, K. H., ADHAM, I. M. and ENGEL, W.Nucleotide sequence of the gene for human transposition protein 1 and its chromosomal localization. Genomics 8(1990)324-330.
20. MALO, D., SCHURR, E., EPSTEIN, D. J., VEKEMANS, M., SKAMENE, E. and GROS, P. The host resistance locus Beg is tightly linked to a group of cytoskeleton-associated protein genes that include villin and desmin. Genomics 10(1991)356-364.
21. MALO, D., VIDAL, S. M., Hu, J., SKAMENE, E. and GROS, P. High-resolution map in the vicinity of the host resistance locus Beg. Genomics 16(1993)655-663.
22. MIYANAGA, K., JUJI, T., MAEDA, H., NAKAJIMA, S. and KOBAYASHI, S. Tuberculoid leprosy and HLA in Japanese. Tissue Antigens 18(1981)331-334.
23. MOHAMED ALI, P. and RAMANUJAM, K. Leprosy in twins. Int. J. Lepr. 34(1966)405-407.
24. NOORDEEN, S. K., LOPEZ BRAVO, L. and SUNDARE-SAN , T. K. Estimated number of leprosy cases in the world. WHO Bull. 70(1992)707-710.
25. O'CONNELL, P., LATHROP, G. M., NAKAMURA, Y., LEPPERT, M. L., LALOUEL, J. M. and WHITE, R.Twenty loci form a continuous linkage map of markers for human chromosome 2. Genomics 5(1989)738-745.
26. OTT, J., Cutting a Gordian knot in the linkage analysis of complex human traits. Invited editorial. Am. J. Hum. Genet. 46(1990)219-221.
27. OTT, J. Analysis of Human Genetic Linkage, (rev. edn.) Baltimore; The Johns Hopkins University Press, 1991.
28. OTTENHOFF, T. H. and DE VRIES, R. R. HLA class II immune response and suppression genes in leprosy. Int. J. Lepr. 55(1987)521-534.
29. OTTENHOFF, T. H., GONZALEZ, N. M., DE VRIES, R. R., CONVIT, J. and VA N ROOD, J.J. Association of HLA specificity LB-E12 (MB 1, DC 1, MT1) with lepromatous leprosy in a Venezuelan population. Tissue Antigens 24(1984)25-29.
30. PRINGEAULT, E., ARPIN, M., GARCIA, A., FINIDORI, J. and LOUVARD, D. A human villin cDNA clone to investigate the differentiation of intestinal and kidney cells in vivo and in cell culture. EMBO 5(1986)3119-3124.
31. RAO, P. S. S., KARAT, A. B. A. and KARAT, S. Epidemiological studies of leprosy in Gudiyatham Taluk. II. Patterns of familial aggregation of leprosy in an endemic area. Lepr. Rev. 40(1969)93-98.
32. RIDLEY, D. S. and JOPLING, W. H. Classification of leprosy according to immunity; a five-group system. Int. J. Lepr. 34(1966)255-273.
33. SAMBROOK, J., FRITSCH, E. F. and MANIATIS, T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press, 1989.
34. SANSONETTI, P. and LAGRANGE, P. H. The immunology of leprosy: speculations on the leprosy spectrum. Rev. Infect. Dis. 3 (1981) 422-469.
35. SCHAUF, V., RYAN, S., SCOLLARD, D., JONASSON, O., BROWN, A., NELSON, K., SMITH, T. and VITHAYASAI , V. Leprosy associated with HLA-DR2 and DQwl in the population of northern Thailand. Tissue Antigens 26(1985)243-247.
36. SCHURR, E., HENTHORN, P. S., HARRIS, H., SKAMENE, E. and GROS, P. Localization of two alkaline phosphatase genes to the proximal region of mouse chromosome 1. Cytogenet. Cell Genet. 52(1989b)65-67.
37. SCHURR, E., SKAMENE, E., FORGET, A. and GROS, P. Linkage analysis of the Beg gene on mouse chromosome 1: identification of a tightly linked marker. J. Immunol. 142(1989)4507-4513.
38. SCHURR, E., SKAMENE, E., MORGAN, K., CHU, M. L. and GROS, P. Mapping of Col3a1 and Col6a3 to proximal murine chromosome 1 identifies conserved linkage of structural protein genes between murine chromosome 1 and human chromosome 2q. Genomics 8(1990)477-486.
39. SHAW, M. A., ATKINSON, S., DOCKRELL, H., HUSSAIN, R., LINS-LAINSON, Z., SHAW, J., RAMOS, F., SILVEIRA, F., MEHDI, S. Q., KAUKAB, F., KHAL-IQ, S., CHIANG, T. and BLACKWELL, J. An RFLP map for 2q33-q37 from multicase mycobacterial and leishmanial disease families: no evidence for an Lsh/Ity/Bcg gene homologue influencing susceptibility to leprosy. Ann. Hum. Genet. 57(1993)251-271.
40. SHIELD, E. D., RUSSEL, D. A. and PERI-CACK-VANCE, M. A. Genetic epidemiology of the susceptibility to leprosy. J. Clin. Invest. 79(1987)1139-1141.
41. SHILOH, Y., DONLON, T., BRUNS, G., BREITMAN, M. L. and Tsui, L.-C. Assignment of the human gamma-crystallin gene cluster (CRYG) to the long arm of chromosome 2, region q33-36. Hum. Genet. 73(1986)17-19.
42. SMITH, D. G. The genetic hypothesis for susceptibility to lepromatous leprosy. Hum. Genet. 50(1979)163-177.
43. SPICKETT, S. G. Genetics and epidemiology of leprosy. I. The incidence of leprosy. Lepr. Rev. 33(1962)76-93.
44. VA N EDEN, W., GONZALEZ, N. M., DE VRIES, R. R., CONVIT, J. and VA N ROOD, J. J. HLA-linkcd control of predisposition to lepromatous leprosy. J. Infect. Dis. 151(1985)9-14.
45. VAN ES, H. H., BOUT, A., Liu, J., ANDERSON, L., DUNCAN, A. M., BOSMA, P., OUDE ELERINK, R., JANSEN, P. L. M., CHOWDHURY, J. R. and SCHURR, E. Assignment of the human UDP glucuronosyltransferasc gene (UGT1A1) to chromosome region 2q37. Cytogenct. Cell. Genet. 63(1993)114-116.
46. VIDAL, S. M., MALO, D., VOGAN, K., SKAMENE, E.and GROS, P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Beg. Cell 73(1993)469-485.
47. VIEGAS PEQUIGNOT, E., LIN, L. Z., DUTRILLAUX, B., APIOUI, F. and PAULIN, D. Assignment of human desmin gene to band 2q35 by a non radioactive in situ hybridization. Hum. Genet. 83(1989)33-36.
48. WAGENER, D. K., SCHAUF, V., NELSON, K. E., SCOLLARD, D., BROWN, A. and SMITH, T. Segregation analysis of leprosy in families of northern Thailand. Gen. Epidemiol. 35(1988)43-52.
49. WEBER, J. NIH/CEPH collaborative mapping group; a comprehensive genetic linkage map of the human genome. Science 258(1992)148-152.
50. WILCOX, E. R., RIVOLTA, M. N., PLOPLIS, B., POTTERF, S. B. and FEX, J. The PAX3 gene is mapped to human chromosome 2 together with a highly informative CA dinucleotidc repeat. Hum. Mol. Genet. 1(1992)215.
1. M.Sc; Institut Territorial de Recherches Médicales Louis Malardc, B. P. 30, Papeete, Tahiti, French Polynesia.
2. Ph.D., Institut Territorial de Recherches Médicales Louis Malardc, B. P. 30, Papeete, Tahiti, French Polynesia.
3. Ph.D., Unite de Génétique Mycobacterienne, (URA CNRS 1300), Institut Pasteur, 25 Rue de Dr. Roux, 75724 Paris 15, France.
4. Ph.D.; McGill Centre for the Study of Host Resistance, Montreal General Hospital Research Institute, Quebec, Canada.
5. M.Sc, McGill Centre for the Study of Host Resistance, Montreal General Hospital Research Institute, Quebec, Canada.
Received for publication on 6 July 1994.
Accepted for publication on 10 August 1994.