- Volume 62 , Number 2
- Page: 229–36
Molecular definition of unique species status of Mycobacterium w; a candidate leprosy vaccine strain
ABSTRACT
Mycobacterium w, a candidate leprosy vaccine strain, is an atypical cultivable mycobacterium. Based on its growth and metabolic properties, M. w was listed in Runyon Group IV, along with other rapid growers such as M. fortuitum, M. smegmatis, M. chelonae and M. vaccae. However, M. w was not fully identical to any one of these. In the present study, a molecular biology approach was used to define the species identity of M. w in a manner that allows reliable comparison to be made with over 30 known mycobacterial species. A 383-bp region, present at the amino terminus of the conserved mycobacterial 65-kDa gene, has been polymerase chain reaction (PCR) amplified in M. w and the DNA sequence was determined. A comparison of the M. w DNA sequence with those of M. tuberculosis, M. avium, M. paratuberculosis and M. fortuitum revealed a species-specific polymorphism, i.e., the presence of nucleotide substitutions unique to M. w. In an alternate approach, a 441-bp region, also a part of the 65-kDa gene, has been PCR amplified in M. w and a Hae III restriction pattern was generated. The 142/127/59-bp Hae III pattern of M. w was found to be unique when compared with M. tuberculosis H37Rv, M. bovis, M. avium, M. intracellulare, M. scrofulaceum, M. kansasii, M. gastri, M. gordonae, M. shimoidei, M. malmoense, M. haemophilum, M. terrae, M. nonchromogenicum, M. triviale, M. marinum, M. flavescens, M. simiae, M. szulgai, M. xenopi, M. asiaticum, M. aurum, M. smegmatis, M. vaccae, M. fortuitum subsp. fortuitum, M. fortuitum subsp. peregrinum, M. chelonae subsp. chelonae, M. chelonae subsp. abscessus and M. genavense; mycobacteria for which the 441-bp Hae III patterns have been documented in the literature. These results established the species identity of M. w at the nucleotide level.RÉSUMÉ
Le Mycobacterium w , une souche candidate pour unvaccin contre la lepre, est une mycobactêrie atypiquecultivable. Sur la base de sa croissance et de ses propriêt Cs mêtaboliques, M. it a etc class& dans le GroupeIV de Runyon, en compagnic d'autres mycobacteriescroissance rapide telles que M. fortuitum, M. smegmails, M. chelonae et M. vaccae . Cependant, M. w n'etait Crement identique a aucun de ceux-ci. Dans l'etude presente, on a utilis une approche de biologie moleculaire pour definir l'identité d'espèce de M. w d'une maniere qui permet de faire une comparaisonfiable avec plus de 30 espèces mycobacteriennesconnues. Une région de 383-bp, presente a l'extremiteamine du gene mycobacterien conserve de 65 kDa, a été amplifiee chez M. w par une reaction de polymeraseen chaine (PCR) et la sequence de l'ADN a etc determince. La comparison de la sequence de l'ADN de M. w avec celle de M. tuberculosis, M. avium, M. paratuberculosis et M.fortuitum a revele un polymorphismespecifique d'espèce, c'est-à-dire la presence de substitutions nucléotidiques uniques a M. w. Dans une approche alternative, une region de 441-bp, faisant ega-lenient partie du gene de 65 kDa, a etc amplifiée par PCR chez M. w et un type de restriction Hae III a été engendre. Le type Hae 111 142/127/59-bp a eté trouvecomme etant unique, par comparaison au H37Rv de M. tuberculosis, M. bovis, M. avium, M. intracellulare,M. scrofulaceum, M. kansaii, M. gastri, M. gordonae, M. shimoidei, M. malmocnse, M. haemophilum, M.terrae, M. nonchromogenicum, M. triviale, M. marl-num, M. Jiavescens, M. simiae, M. sulgai, M. xenopi,M. asiaticum, M. aurun:, M. smegmatis, M. vaccae, M. fortuitum subsp.fin-tuiturn, M. fortuitum subsp. peregrinum, M. chelonae subsp. chelonae, M. chelonae subsp. abscesses et M. genavense , mycobacteries pourlesquelles le type Hae Ill 441-bp a été documenté dansla litterature. Ces résultats etablissent l'identite d'espèce de M. w au niveau du nucléotide.RESUMEN
Mycobacterium w es una micobacteria atípica cultivable que se ha considerado como candidato para preparar una vacuna contra la lepra. Con base en su crecimiento y propiedades metabólicas, M. w se clasificó en el grupo Runyon IV, junto con otras micobacterias de crecimiento rápido tales como M. fortuitum, M. smegmatis, M. chelonae, y M. vaccae. Sin embargo, M. w no fue completamente idéntica a ninguna de ellas. En el presente estudio se aplicaron las técnicas de la biología molecular para definir la identidad de especie de M. w por comparación con más de 30 especies micobacterianas conocidas. Para ésto, se amplificó por la reacción en cadena de la polimerasa (PCR) una región de 383 pb del gene que codifica la proteína altamente conservada de 65 kDa (extremo amino terminal) de las micobacterias. La secuenciación del DNA amplificado, y la comparación de esta secuencia con aquellas correspondientes a M. tuberculosis, M. avium, M. paratuberculosis, y M. fortuitum, revelaron un polimorfismo específico de especie, esto es, la presencia de substituciones de nucleótidos únicos de M. w. En un enfoque alternativo, una región de 441 pb, también una parte del gene de 65 kDa, fue amplificada por PCR en M. w para establecer su patrón de restricción con Hac III. El patrón Hae III de 142/127/ 59-pb de M. w resultó único cuando se comparó con el patrón de restricción correspondiente a M. tuberculosis H37Rv, M. bovis, M. avium, M. intracellularc, M. scrofulaceum, AI. kansasii, M. gastri. M. gordonae, M. shimoidei, M. malmoense, M. haemophilum. M. terrae. M. nonchromogenicum, M. triviale, M. marinum, M. flavescens, M. simiae, M. szulgai, M. xenopi, M. asiaticum, M. aurum, M. smegmatis, M. vaccae, M. fortuitum subsp. fortuitum. M. fortuitum supsp. peregrinum, M. chelonae subsp. chelonae. M. chelonae subsp. abscessus, and M. genavense, micobacterias para las cuales los patrones de resetricción 441 pb Hae III han sido documentados en la literatura. Estos resultados establecen la identidad de especie de M. w a nivel de nulceótidos.In the course of investigations carried out to select a mycobacterium as a candidate vaccine for leprosy, a number of known species of mycobacteria and several atypical strains were examined (22). Their ability to generate blast transformation and the production of macrophage migration inhibition factor(12) similar to that elicited by Mycobacterium leprae from peripheral blood leukocytes of a panel of polar tuberculoid leprosy (TT) patients was determined. The "preferred" mycobacteria in this test (under code names) were subsequently screened for immunogenicity (14) and generation of delayed-type hypersensitivity (DTH) responses to homologous antigens and M. leprae (3, l3). Finally, autoclaved suspensions of five coded mycobacteria were employed for Dharmendra and Mitsuda tests in TT and lepromatous leprosy (LL) patients. Mycobacterium w elicited DTH responses not only in TT but also in LL patients who do not normally respond to M. leprae lepromin (5. 6, 8, 15, 17), shares both B- (4) and T-cell (11, 26) epitopes with M. leprae, but has additional determinants overriding the nonresponsiveness to M. leprae seen in LL patients.
Autoclaved suspensions of M. w (5 x 108 bacilli in 0.1 ml saline) have been evaluated as a vaccine. Phase II/III immunotherapeutic trials with this vaccine in multibacillary, lepromin-negative leprosy patients have shown that the administration of the vaccine every 3 months to the patients treated with standard multidrug therapy (MDT) caused a significantly accelerated clearance of the bacilli and clinical improvement (23,24,27) over 80% of the vaccinated patients became lepromin positive. The period of time to release from treatment was significantly shortened (27). The vaccine was also effective in patients who were slow responders or nonresponders to drugs.
Histopathologically, many patients demonstrated upgrading and clearance of dermal granuloma to a nonspecific infiltration (NSI) status (10). The M. w vaccine is currently in large scale Phase III/IV immunotherapeutic-cum-immunoprophylactic trials in District Kanpur Dehat of North India, and is also an arm of the comparative immunoprophylactic trials with the World Health Organization (WHO) vaccine in South India. No serious ill effects due to the administration of the vaccine have been reported.
This communication enquires on the species to which M. w belongs. Its growth and metabolic properties resemble those of mycobacteria listed in Runyon's Group IV (9,19). However, it is not fully identical to any one of these on the basis of one or the other properties. The present study was undertaken to establish the identity of M. w by employing molecular biology techniques. Hance, et al. have proposed the identification of mycobacteria from the base sequence of a polymorphic region of the mycobacterial 65-kDa gene (7). Telenti, et al. have employed a PCR-restriction enzyme pattern analysis (PRA) method for identification to the species level of mycobacteria 25). These techniques were employed by us to determine the species identity of M. w.
MATERIALS AND METHODS
Mycobacterial strains. M. tuberculosis and M. w were grown in Middlebrook 7H9 medium. Genomic DNA from mycobacteria was isolated by the method described earlier (16).
Oligonucleotides. Oligonucleotides TBI (5'GAGATCGAGCTGGAGGATCC) and TB2 (5'AGCTGCAGCCCAAAGGTGTT) have been described by Hance, et al. (7).TB11 (5'ACCAACGATGGTGTGTCCAT) and TB12 (5'CTTGTCGAACCGCATA-CCCT) were reported by Telenti, (25).Amplification of mycobacterial DNA witha) TB I and TB2 resulted in a 383-bp fragment (corresponding to 423-805-bp regionof the M. tuberculosis 65-kDa gene, and b)TB I 1 and TB12 resulted in a 441-bp fragment (corresponding to 396-836-bp region of the M. tuberculosis 65-kDa gene (21). Oligonucleotides were synthesized by the phosphoramidite method.
PCR amplification. Genomic DNA from M. tuberculosis or M.w was PCR amplified(18) using TB1 + TB2 or TB11 + TB12primer combinations. The composition ofthe PCR mixture (100 µl) was 10 mM Tris-HCI at pH 8.3, 50 mM KCI, 1.5 mM Mg Cl2,0.01% (w/v) gelatin, 200 µM each of deox-ynucleotide triphosphates (dNTP), 20 µM each of primers, 50 ng of genomic DNA,and 2.5 units of Taq DNA polymerase.Thirty cycles of amplification were per-formed. Each cycle included denaturationat 94°C for 1 min, annealing at 55°C for 2min, and extension at 72°C for 2 min.
DNA sequencing. The 383-bp PCR-amplified product of M. w (using primers TBI + TB2) was ethanol precipitated and resuspended in TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH 8.0). Primers TB1 and TB2 contained Bam HI and Pst I sites, respectively. The PCR product was cleaved with restriction cndonucleases Bam HI and Pst I as per manufacturer's instructions (Promega, Madison, Wisconsin, U.S.A.) and cloned into M13 mpl8 and M13 mpl9 (BRL, Bethesda, Maryland, U.S.A.). Six independent M13 clones bearing the M. w 383bp fragment in either orientation have been sequenced by Sanger's dideoxy method as per manufacturer's instructions (Taq Track; Promega).
PCR-restriction enzyme pattern analysis (PRA). The 441-bp PCR-amplification products of M. tuberculosis and M. w genomic DNA using primers TB 11 and TB 12 were ethanol precipitated and resuspended in TE buffer. An aliquot of each was then digested with restriction endonuclease Hae III, as per the manufacturer's instructions (Promega). The undigested and Hae III digested samples were electrophoresed through a 3% agarose (BRL) gel.
Elimination of PCR contamination. In order to avoid template DNA or PCR product contamination, the PCR amplification reactions were mixed in a separate room. Each PCR experiment included the negative control, i.e., no template DNA added to the reaction mixture. There was consistently no amplication in this tube, which ruled out aerosol or reagent contamination. The post-PCR DNA manipulations, such as gel electrophoresis and cloning procedures, were performed in a room away from where PCR reactions are mixed. We found this to be efficient in avoiding PCR contamination.
RESULTS AND DISCUSSION
In order to define the species status of M. w at the nucleotide sequence level, we undertook two separate approaches, both of which have been shown to be reliable for mycobacterial species identification:
A 383-bp sequence situated at the amino terminus in the open reading frame (423805-bp) of the gene coding for 65-kDa mycobacterial antigen (21) has been shown to be conserved among several species of mycobacteria. By PCR amplification and DNA sequencing, Brisson Noel, et al. observed species-specific polymorphism at the nucleotide level within this region (7). The 383bp region was PCR amplified from the M. w genomic DNA by using primers TBI and TB2, subcloned in both orientations and nucleotide sequence determined as described in Materials and Methods. A comparison of the 383-bp sequences of M. w and other mycobacteria reported so far showed considerable homology, as is expected of a conserved gene. Significantly, there were also polymorphic nucleotide substitutions in M. w. The nucleotide sequence of the amplified region of M. w differed from that of M. tuberculosis at 24 out of 343 bases (Fig. 1A). It is interesting to note that 20 of these substitutions did not alter the amino-acid sequence (Fig. IB), thereby reflecting the conserved nature of the 65-kDa heat-shock protein. However, changes in the nucleotide sequence at positions 206-208 and at 232 resulted in amino-acid substitutions with reference to M. tuberculosis (Fig. 1, italics). The M. w sequence also differed considerably from that of M. avium, M. paratuberculosis and M. fortuitum particularly between nucleotides 249 and 266, a region shown to be highly polymorphic by Brisson Noel, et al. (7). More importantly, when compared to the sequences of M. tuberculosis, M. avium, M. paratuberculosis and M. fortuitum, M. w exhibited the presence of unique nucleotide substitutions at positions 94, 121, 130 and 286 bp (Fig. 1A, bold letters), which could be termed as species-specific to M. w with reference to the mycobacterial sequences available in literature.

Fig. 1. A = The DNA sequence of 343-bp fragment (excluding the sequence of the primers TBI and TB2 from the amplified 383-bp fragment) of the AT. W 65-kDa gene was aligned with that of AT. TUBERCULOSIS (AT. TB). The symbol (-) indicates identity of bases with the sequence shown for AT. TB. Nucleotide substitutions are indicated by the altered base. Base substitutions that gave rise to new codons in AL. W are shown in italics. In addition to AT. TB, the 343-bp sequence of A/,W also was compared with those of AT. AVIUM, AT. PARATUBERCULOSIS and AT. FORTUITUM, which are available in the literature (7). Such an analysis indicated that the four nucleotides at positions 94, 121, 130 and 286 bp (shown in bold) are unique to AT. W (the first nucleotide after the primer TBI is numbered as 1, and the last nucleotide before the TB2 primer starts is numbered as 343). B = A comparison of deduced amino-acid sequence of the 343-bp region of M. TB and AT. W shown in A. The (-) symbol indicates identical amino acids in the AT. W protein with reference to AT. TB. Altered amino acids are indicated by the one letter code in AT. W sequence. It is noteworthy that although there were as many as 24 nucleotide substitutions in AF. W (A), 20 of them were silent mutations. Only substitution of GGC (AT. TB) to TCG (AT. W) has changed the glycine to a serine, GAG (AT. TB) to GAC (A/, W) resulted in a change from glutamic acid to aspartic acid (italics).
Although the features within the 383-bp sequence demonstrated the uniqueness of M. w, comparison is limited to only a few mycobacteria in which this gene has been sequenced. Telenti, et al. (25) have recently reported a PCR-restriction enzyme pattern analysis (PRA) method which allows comparison to be made among a large number of mycobacteria for identification to the species level without the need for DNA sequencing. In this method, a 441-bp fragment of the 65-kDa gene (nucleotides 396 through 836 of the M. tuberculosis 65-kDa gene) was amplified, and the resultant amplicon was digested separately with restriction enzymes list EII and Hac III. It was observed that the restriction fragment patterns derived in this manner were distinctive for each mycobacterial species. Based on this observation Telenti, et al. compiled a database of Bst EII and Hae III fragment patterns for a large number of mycobacterial species (25). A comparison of the Bst EII or Hae III restriction fragment pattern of the 441-bp region of an unknown mycobacterium with the known patterns in the databank should then reveal the identity of the unknown to a species level.
We chose to apply this method to investigate M. w identity for two reasons: a) The restriction fragment pattern of the M. w -441bp region can be generated and documented for future reference, and comparison can be made with a large number of mycobacterial species available in the database of Telenti, et al. (25); b) Knowledge of the nucleotide sequence of all but a few of the 441 bp of M. w (as depicted in Fig. 2A) permits determination of the size of restriction fragments with an accuracy that could match the computer-aided list in the databank (25).
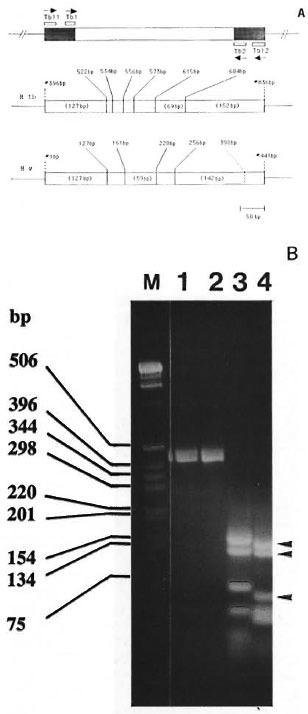
Fig. 2. A = Schematic representation of the region of the 65-kDa gene analyzed in this study. Top panel shows the location of the PCR primers and direction of DNA synthesis (arrows). The 343-bp segment between primers TBI and TB2 has been sequenced in At. w (Fig. 1A). The 441-bp fragment, amplified by TBI 1 and TBI2, was used for generating the Hae III restriction pattern. The shaded area represents the part where the DNA sequence is unknown in At. w. In At. tuberculosis (At. tb), TB11 and TB12 amplified the 441 bp segment between positions #396 bp and #836 bp of the 65-kDa gene (2I). The middle panel in the figure shows the location of Hac III sites within the 441-bp region of At. tb (numbering of the base pairs is done according to Shinnick21). The length of the major Hae III fragments is indicated in parentheses. The lower panel shows the 441-bp fragment of M. w, indicating the known Hae III sites. As discussed in the text, at position #398 bp. within the region corresponding to the location of TB2 primer, a silent mutation possibly has created an additional Hae 111 site (shown by dashed line). The major At. w Hae III fragments 142/127/59 (seen in Fig. 2B, lane 4) are indicated in parentheses.
B = Genomic DNA from At. tuberculosis (lane 1) and At. w (lane 2) was amplified using primers TBI 1 and TBI2. The resultant 441-bp band (lanes 1 and 2) was digested with Hac III and separated by 3% agarose gel electrophoresis, which is shown in lanes 3 and 4 for At. tuberculosis and At. w, respectively. The 142-127-59-bp Hae III fragments of At. w are indicated by the arrowheads. Lane M contains the 1-kb-size markers listed on the left.
To serve as a standard for PCR amplification and restriction fragment sizing, we have included M. tuberculosis for which the complete sequence of the 65-kDa is known M. w and M. tuberculosis genomic DNAs were amplified by TBI 1 and TBI2, which resulted in the amplification of the expected 441-bp band in both species (Fig. 2B, lanes 1 and 2). The amplicons were then digested with Hae III and the restriction pattern was generated by agarose gel electrophoresis. M. tuberculosis produced Hae III bands of 152, 127 and 69 bp in size (Fig. 2B, lane 3), which is in agreement with the expected pattern (127, 12, 22, 17, 42, 69, and 152 bp) deduced from the published sequence (fragments smaller than 50 bp are not well resolved in Fig. 2B). This indicated that the conditions used for amplification and restriction endonuclease cleavage were correct. Based on the presence of Hae III sites in the portion of 441-bp band of M. W for which DNA sequence is known (Figs. 1 and 2), the expected bands are 127, 34, 59, 36 and 185 bp, of which the 127- and 59bp bands can be seen in Fig. 2B, lane 4. The expected 185-bp band (256-441 bp) is missing and, instead, a 142-bp band is present (Fig. 2B, shallow arrow), indicating the presence of a Hae III site in the part of 441 bp for which sequence is unknown. The following is a possible explanation.
Within the oligonucleotide sequence TB2 (this forms the 392-412-bp part of the 441 bp fragment as depicted in Fig. 2), which is derived from M. bovis BCG 65-kDa gene, there is a GGGC sequence. With respect to the reading frame of the 65-kDa gene, GGG form a codon that codes for glycine, while the "C" is part of the next codon CTG. From a comparison of the 343-bp sequence (Fig. 1A), it was noted that nucleotide substitutions occurred in this gene predominantly in the third base position of a codon such that the reading frame and the resulting amino acid are not altered. It is possible then, that the GGG of M. bovis BCG within the TB2 sequence has changed to GGC in M. w , which would still code for the same amino acid (glycine) but, more importantly, has now given rise to GGCC and thus created an additional Hae III recognition site. As a result of this, the expected 185-bp band was cleaved to yield a 142-bp (Fig. 2B, lane 4, arrow) and a 43-bp fragment (because of the presence of several fragments in this size range, resolution of 43-bp fragment is poor in Fig. 2B, lane 4). The M. w Hae III pattern generated by the PRA method is, therefore, 142/127/59 bp. A comparison of the M. w Hae III PRA profile with that of several other mycobacterial species (listed below) included in the databank (25) clearly indicated that this banding pattern is unique to M. w and is not represented by any other mycobacterial species tested.
The results from the two different experiments, i.e., DNA sequencing of the 383-bp fragment and restriction fragment pattern analysis of the 441-bp fragment, both of which are a part of the conserved mycobacterial 65-kDa gene, allowed either a nucleotide sequence or a PRA pattern comparison to be made with M. tuberculosis H37Rv, M. bovis, M. bovis BCG, M. avium, M. intracellulare, M. scrofulaceum, M. paratuberculosis, M. kansasii, M. gastri, M. gordonae, M. shimoidei, M. malmocnse, M. haemophilum, M. terrae, M. nonchromogenicum, M. triviale, M. marinum, M. flavescens, M. simiae, M. szulgai, M. xenopi, M. asiaticum, M. aurum, M. smegmatis, M. vaccae, M. fortuitum subsp. fortuitum, M. fortuitum subsp. peregrinum, M. chelonae subsp. chelonae, M. chelonae subsp. abscessus and M. genavense, which clearly showed that M. w is a species that differs from these mycobacteria.
The analysis of nucleotide sequence of conserved genes already has been shown to be useful in the identification of mycobacteria to a species level. Recently, an isolate of the genus Mycobacterium derived from HIV-infected patients has been identified as new, and assigned a species name M. genavense based on the observation that the ribosomal RNA sequence of this isolate showed specific nucleotide substitutions different from previously known mycobacterial species (2). The Bst EII and Hae III restriction patterns of the 441-bp fragment of M. genavense have also shown this organism to be a new species, which confirmed the merits of the PRA method in determining species status of an unknown mycobacterium (25). Like the 65-kDa gene in our study, 16S rRNA genes also have been sequenced in several mycobacteria, and hypervariable regions within the gene were identified containing signature sequences at the species level (1). Potentially, rRNA gene sequencing also could be used as an approach to substantiate the unique species status of M. w.
In summary, the molecular biology approach used in this study has shown M. w to be a unique species, which is in agreement with our earlier characterizations by traditional methods (9, 19).
Acknowledgment. This work was supported by the Department of Biotechnology, Government of India.
REFERENCES
1. BODDINGHAUS, B., ROGALL, T., FLOHR, T.,BLOCKER, H. and BOTrGER, E. C. Detection andidentification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28(1990)1751-1759.
2. BOTTGER, E., TESKE, A., KRISCHNER, P., BOST, S.,CHANG, H., BEER, V. and HIRSCHEL, B. Disseminated " M. genavense " infection in patients withAIDS. Lancet 340(1992)76-80.
3. FOTEDAR, A., MEHRA, N. K., MUSTAFA, A. S. and TALWAR, G. P. Local reactions to intradermalinstillation of Mycobacterium w and ICRC bacilliin mice. Lepr. India 50(1978)520-533.
4. GANJU, L., MUKHERJEE, R., BATRA, H. V. and TALWAR, G. P. Immunoblot analysis of antigens of Mycobacterium w. A candidate antileprosy vaccine using monoclonal antibodies and patient sera.Int. J. Med. Microbiol. 273(1990)378-385.
5. GIRDHAR, B. K. and DESIKAN, K. V. Results of skin tests with five different mycobacteria. Lepr.India 50(1978)555-559.
6. Goya., D. C. and BHUTANI, L. K. Delayed hypersensitivity skin reactions to lepromin and antigens prepared from four other mycobacteria. Lepr.India 50(1978)550-554.
7. HANCE, A., GRANDCHAMP, B., LEVY-FREBAULT, V.,LECOSSIER, D., RAUZIER, J., BOCART, D. and GICQUEL, B. Detection and identification of mycobacteria by amplification of mycobacterial DNA.Mol. Microbiol. 3(1989)843-849.
8. HOGERZEIL, L. M. and PRABHUDASS, N. Delayed hypersensitivity skin reactions to lepromins prepared from M. leprae and selected cultivable mycobacteria. Lepr. India 50(1978)560-565.
9. KATOCH, V. M. A report on the biochemical analysis of Mycobacterium w. Lepr. India 53(1981)385-389.
10. MUKHERJEE, A., ZAHEER, S. A., SHARMA, A. K.,MISRA, R. S., KAR, H. K., MUKHERJEE, R. and TALWAR, G. P. Histopathological monitoring of immunotherapeutic trials with Mycobacterium w . Int. J. Lepr. 60(1992)28-35.
11. MUSTAFA, A. S. Identification of T-cell activating recombinant antigens shared among three candidate antileprosy vaccines, killed M leprae, A1. BCG and Alveobacterium w. Int. J. Lepr. 56(1988)265-273.
12. MUSTAFA, A. S. and TALWAR, G. P. Five cultivable mycobacterial strains giving blast transformations and leukocyte migrationinhibition ofleukocytes analogous to Mycobacterium leprae . Lepr. India 50(1978)498-508.
13. MUSTAFA, A. S. and TALWAR, G. P. Delayed hypersensitivity skin reactions to homologous and heterologous antigens in guinea pigs immunized with M. leprae and four selected cultivable mycobacterial strains. Lepr. India 50(1978)509-519.
14. MUSTAFA, A. S. and TALWAR, G. P. Enlargementof draining lymph nodes in mice by four selected cultivable strains of mycobacteria. Lepr. India 50(1978)534-538.
15. MUSTAFA, A. S. and TALWAR, G. P. Early and latereactions in tuberculoid and lepromatous leprosy patients with lepromins from Mycobacterium leprae and five selected cultivable mycobacteria. Lepr.India 50(1978)566-571.
16. REDDI, P. P., TALWAR, G. P. and KHANDEKAR, P.S. Repetitive DNA sequences of Mycobacterium tuberculosis: analysis of differential hybridizationpattern with other mycobacteria. Int. J. Lepr. 56(1988)592-597.
17. SAHIB, H. S. M. and VELLUT, C. Some observations on skin reactions by lepromin and four other mycobacterial antigens. Lepr. India 50(1978)579-587.
18. SAIKI, R. K., GELFAND, G. H., STOFFEL, S., SCHARF,S. J., HIGUCHI, R., HORN, G. T., Mullis, K. B.and ERLICH, H. A. Primer-directed enzymaticamplification of DNA with a thermostable DNA polymerase. Science 239(1988)487-491.
19. SAXENA, V. K., SINGH, U. S. and SINGH, A. K. Bacteriological study of a rapidly growing strainof mycobacterium. Lepr. India 50(1978)588-596.
20. SHARMA, R. C. and SINGH, R. Comparative study of skin reactions in leprosy patients to M. leprae lepromin and antigens from cultivable mycobacteria. Lepr. India 50(1978)572-578.
21. SHINNICK, T. The 65-kilodalton antigen of Mycobacterium tuberculosis. J. Bacteriol. 169(1987)1080-1088.
22. TALWAR, G. P. Towards development of a vaccine against leprosy. Lepr. India 50(1978)492-497.
23. TALWAR, G. P., ZAHEER, S. A., MUKHERJEE, R.,WALIA, R., MISRA, R. S., SHARMA, A. K., KAR, H.K., MUKHERJEE, A., PARIDA, S. K., SURESH, N. R.,NAIR, S. K. and PANDEY, R. M. Immunotherapeutic effects of a vaccine based on a saprophyticcultivable mycobacterium, Mycobacterium w , inmultibacillary leprosy patients. Vaccine 8(1990)121-129.
24. TALWAR, G. P., ZAHEER, S. A., SURESH, N. R.,PARIDA, S. K., MUKHERJEE, R., SINGH, 1. G., SHAR-MA, A. K., KAR, H. K., MISRA, R. S. and MUKIIER-JEE, A. Immunotherapeutic trials with a candidateanti leprosy vaccine based on Mycobacterium w .Trop. Med. Parasitol. 41(1990)369-370.
25. TELENTI, A., MARCHESI, F., BALZ, M., BALLY, F.,BoTTGER, E. C. and BODMER, T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzymeanalysis. J. Clin. Microbiol. 31(1993)175-178.
26. YADAVA, A., SURESH, N. R., ZAHEER, S. A., TAL-WAR, G. P. and MUKHERJEE, R. T cell responsesto fractionated antigens of ,Mycobacteriton w, acandidate anti leprosy vaccine, in leprosy patients.Scand. J. Immunol. 34(1991)23-31.
27. ZAHEER, S. A., MUKHERJEE, R., RAM KUMAR, B.,MISRA, R. S., SHARMA, A. K., KAR, H. K., KAUR,H., NAIR, S. K., MUKHERJEE, A. and TALWAR, G.P. Combined multidrug and Mycobacterium w vaccine therapy in patients with multibacillaryleprosy. J. Infect. Dis. 167(1993)401-410.
1. Ph.D.; National Institute of Immunology, New Delhi 110067, India.
2. M.Sc; National Institute of Immunology, New Delhi 110067, India.
3. Ph.D.; National Institute of Immunology, New Delhi 110067, India.
4. D.Sc., National Institute of Immunology, New Delhi 110067, India.
Received for publication on 9 November 1993;
Accepted for publication in revised form on 9 March 1994.