- Volume 61 , Number 4
- Page: 600–4
A simple and rapid technique for the detection of rifampin resistance in Mycobacterium leprae
ABSTRACT
The rifampin resistance of Mycobacterium leprae is due to missense mutations in the rpoB gene encoding the β-subunit of the essential enzyme RNA polymerase. A rapid and very simple method has been developed to detect rifampin resistance in small numbers of M. leprae present in biopsies. It involves polymerase chain reaction amplification of a defined region of the rpoB gene followed by single-strand conformational polymorphism analysis (PCR-SSCP). The reliability of the method has been tested on a sample of known drug-resistant and susceptible isolates of M. leprae.RÉSUMÉ
La résistance à la rifampicine de Mycobacterium leprae est due à des mutations désordonnées dans le gène rpoli encodant la sub-unité β de l'enzyme essentiel ARN-Polymerase. Une méthode rapide et très simple at été développée pour détecter la résistance à la rifampicine sur les faibles qualities de M. leprae présentes dans les biopsies. Elle inclut la réaction d'amplification de la polymerase en chaîne d'une région précise du gène rpoB suivie d'une analyse polymorphique de la conformation d'un brin momocaténairc (PCR-SSCP). La fiabilité de la méthode a été testée sur un échantillon d'isolates de M. leprae dont la résistance ou la sensibilité à la rifampicine était connue.RESUMEN
La resistencia del Mycobacterium leprae a la rifampina se debe a mutaciones sin sentido en el gene rpoB que codifica la subunidad β de la enzima esencial RNA-polimerasa. Nosotros desarrollamos un método rápido y muy simple para detectar la resistencia a rifampina en pequeños números de M. leprae presentes en biopsias. El método involucra la amplificación por la reacción en cadena de la polimerasa de una región definida del gene rpoB, seguido por un análisis del polimorfismo conformational del DNA de cadena simple (PCR-SSCP). La confiabilidad del método se lia probado usando especímenes de M. leprae susceptibles y resistentes a la droga.Rifampin is the key component of the standard multidrug regimen used for the treatment of leprosy (5,13). It would be highly desirable to have at our disposal a rapid, yet simple, technique for monitoring suspected cases of rifampin resistance which could be applied directly to biopsy material. This could replace the extremely long rifampin-susceptibility test, currently performed after inoculation of mice (11), or the radiorespirometric method which requires relatively large numbers of metabolically active Mycobacterium leprae (2,3). In a recent study, the molecular basis of rifampin resistance in M. leprae was established (7) by direct DNA sequence analysis of the rpoB genes, amplified by the polymerase chain reaction (PCR) (14) from drug-resistant isolates. Resistance was shown to result from a limited number of missense mutations located within a short stretch of the gene. Since direct sequencing of PCR products is a relatively laborious approach, the single-strand conformational polymorphism (SSCP) technique developed by Orita (10) for detecting point mutations and other polymorphisms has been adopted. When applied to the mutants characterized previously (4,7), the M. leprae isolates could be classified as rifampin-resistant or -sensitive in less than 2 days. The method has been appraised by analyzing a new, putativcly rifampin-resistant mutant of M. leprae isolated from a treated lepromatous leprosy patient who had relapsed.
MATERIALS AND METHODS
M. leprae strains. The nine rifampin-resistant mutants characterized in detail by Grosset, et al. (4) and analyzed recently by DNA sequencing (7) were again employed together with the five rifampin-susceptible isolates used previously as controls (The Table). A new strain, 92041 (The Table), was detected in biopsy material obtained from a lepromatous leprosy patient originally from Martinique but now resident in France. The patient had been treated with dapsone monotherapy from 1958-1977, and then with rifampin alone for 2 years before starting a course of rifampin plus prothionamide. The patient, who appears to have taken his medication irregularly, relapsed after some years and was suspected of harboring rifampin-resistant bacilli.
PCR procedures. The region of rpoB known to harbor mutations conferring rifampin resistance was amplified directly from "freeze-boiled" biopsies (14) by the polymerase chain reaction (PCR) using primers Brpo22 (CAGGACGTCGAGG-CGATCAC) and rpo32 (TCCTCGTCAG-CGGTCAAGTA), as described previously (7). This gave rise to a fragment of 390 bp which was uniformlv labeled with 32P by including 5 µCi of (α32P) dCTP (3000 Ci/ mmole; Amersham International, Amersham, U.K.) in the PCR reaction which was performed using 35 cycles (1 min at 92ºC, 2 min at 61ºC and 2 min at 72ºC) with a final elongation time of 10 min at 72ºC. DNA sequences were obtained directly from unlabeled PCR products as described (7).
SSCP analysis. To detect mutations in the rpoB fragment, single-strand conformation polymorphism (SSCP) analysis was employed using the conditions described recently (8,12). Briefly, 10 µl of the 32P-labeled PCR fragment was mixed with 15 µl of H2O and 25 µl of a solution containing 0.1% sodium dodecyl-sulfate (SDS) and 2 raM ethylene-diamine tetra-acetic acid (EDTA). A 5 µl aliquot was mixed with sample buffer (95% formamide, 20 raM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) and heated at 95ºC for 10 min to denature the PCR fragment. Samples were quickchilled on ice to prevent renaturation and immediately loaded onto a 5% polyacrylamide gel (30% acrylamide/0.5% bis-acrylamide; 20 x 50 cm) containing TBE buffer (89 raM Tris, 89 mM boric acid, 2 raM EDTA). Electrophoresis was conducted for 2.5 hr at constant power (65W) using the TBE buffer. The gel was dried and subjected to autoradiography.
In some experiments, the size of the PCR fragment was reduced by digestion with restriction endonuclease Pvul (10 µl PCR reaction, Pvul buffer, 4 units of enzyme in a final volume of 25 µl) for 1 hr, thus yielding two fragments of 239 bp and 149 bp (the larger of which carries the mutations). This procedure enhanced the resolution of the strands on the gel and accentuated the differences due to the mutations. When the digestion was performed, samples were mixed directly with an equal volume of a solution containing SDS and EDTA and then processed as described.
RESULTS
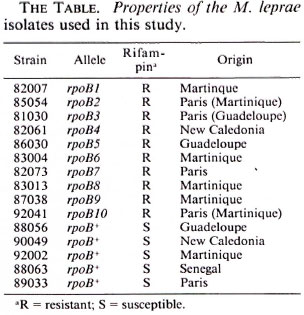
It has been shown recently in nine independent isolates of M. leprae, that rifampin resistance is due to tightly clustered mutations in a short region of the rpoB gene (7). In seven cases, a single base change was found by DNA sequencing; in the eighth mutant two consecutive nucleotides deviated from the wild type rpoB sequence (6,7). A 6 bp insertion was detected in the remaining mutant. These findings are summarized in Figure 1.
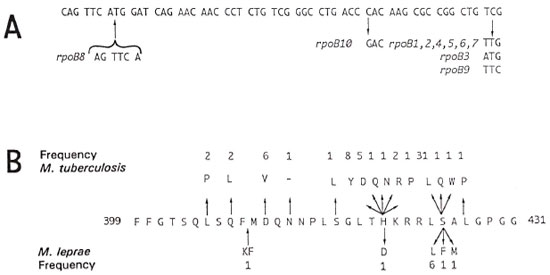
Fig. 1. A. Nucleotide sequence of a short region of rpoB from M. leprae harboring mutations conferring rifampin resistance. Base changes and corresponding alleles are indicated. B. Amino-acid sequence comparison of part of region II, of the β subunit of RNA polymerase from M. tuberculosis and M. leprae. Residue numbers are indicated together with the amino acids in the one letter code. Mutated amino-acid residues associated with rifampin resistance are shown along with the frequency with which a given mutation has been isolated. Data for M. tuberculosis were taken from (8,12). One letter codes for common amino acids are as follows: A = alanine; C = cysteine; D = aspartic acid; F = phenylalanine; G = glycine; H = histidinc; K = lysine; L = leucine; M = methionine; N = asparagine; P = proline; Q = glutamine; R = arginine; S = serine; T = threonine; V = valine; W = tryptophan; Y = tyrosine.
Since DNA sequencing is relatively timeconsuming and labor-intensive, we decided to adapt the SSCP technique (10) in order to develop a simpler means of detecting mutations. Consequently, PCR was employed to amplify the region of rpoB, which has been found to be prone to mutation, from the nine resistant mutants studied previously and five susceptible strains. In addition, to test the power of the method, acidfast bacilli (AFB) harvested from the skin biopsy of a previously treated patient (92041; The Table), who had relapsed after rifampin therapy, was included in the PCR-SSCP analysis. Initially, the migration of the strands from the 390 bp PCR fragment obtained from the rpoB gene of 15 different strains was examined by electrophoresis. Although the differences in the mobilities of the strands were suboptimal, striking differences were apparent (Fig. 2). All of the rifampin-sensitive strains displayed the same electrophoretic pattern; the electrophoretic patterns of the mutants differed significantly. The mobilities of the strands from strains harboring the same mutations were identical but those from the other mutants, including the bacilli from the biopsy from patient 92041, displayed another pattern (Fig. 2A).
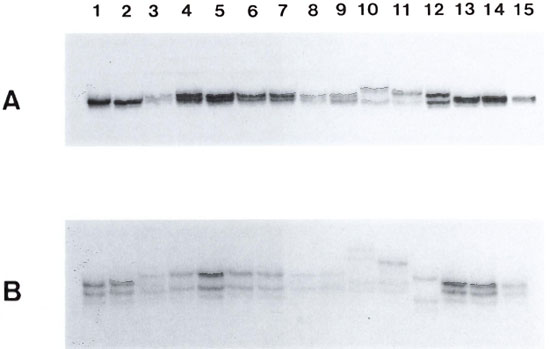
Fig. 2. A. SSCP analysis of the 390 bp PCR fragment obtained from the rpoB genes of various rifampinsensitive (S) and -resistant (R) M. leprae strains. Lane I, 88056 (S); lane 2, 90049 (S); lane 3, 82007 (R); lane 4, 85054 (R); lane 5, 86030 (R); lane 6, 82061 (R); lane 7. 86030 (R); lane 8, 83004 (R); lane 9, 82073 (R); lane 10 83013 (R); lane II. 87038 (R); lane 12, 92041 (R); lane 13, 92002 (S); lane 14, 88063 (S); lane 15, 89033 (S). B. SSCP analysis of the 249 bp fragment obtained on PvuI digestion of the 390 bp PCR fragment obtained from the rpoB genes of various M. leprae strains. Samples are as described in Figure 2A.
To increase the resolution of the technique, the size of the PCR fragment was reduced by cleavage with PvuI. After SSCP-electrophoresis, the differences in the mobilities of the strands were much more obvious (Fig. 2B). Since the electrophoretic pattern obtained with the bacteria isolated from patient 92041 differed from that of the drug-sensitive strains this suggested that its rpoB gene, allele rpoB10, was indeed mutated. Furthermore, it also differed from all of the other mutants, thus indicating that a new type of mutation was present. To confirm this hypothesis the PCR fragment was subjected to DNA sequence analysis (data not shown). A single base change, C to G (Fig. 1 A) from the wild type sequence (6,7), was found in the 200 bp segment examined and this would lead to the replacement of His-520 by Asp in the β-subunit of RNA polymerase (Fig. IB).
DISCUSSION
SSCP analysis is an extremely powerful technique for detecting mutations in short DNA fragments, and has found great application in the field of human genetic diseases such as cystic fibrosis or phenylketonuria (1,10). In the present study, we have shown that it is also a most useful tool for screening for rifampin resistance in M. leprae. Initially, the method was optimized by using a panel of well-characterized resistant and susceptible strains, and striking differences in the electrophoretic mobilities of the strands of wild-type and mutant alleles of rpoB were seen. In this work PCR fragments were labeled with 32P for reasons of ease. For routine application several alternatives are available, including the use of fluorescently tagged primers, incorporation of easily detectable modified bases, such as digitonin-labeled dUTP, or silver staining of unlabeled DNA (1,9). Further improvements could be obtained by using primers, flanking the region of interest, that give rise to a smaller PCR product, thus obviating the need for the PvuI digestion. To test the predictive powers of SSCP, bacilli suspected of being rifampin resistant, from a skin biopsy of patient 92041, were analyzed and a novel electrophoretic pattern was found. Subsequent DNA sequence studies revealed a C-to-G transversion in codon 420 of rpoli which resulted in the substitution of a histidine residue by aspartic acid. Exactly the same mutation has been found in rifampin-resistant isolates of M. tuberculosis (Fig. 1) (8,13) thus indicating that bacilli from biopsy 92401 almost certainly are resistant. To confirm this, their drug susceptibility is currently being assessed in mice although the results will not be available for 1 year. This point underlines the great value of a combined PCR-SSCP approach because one can perform the reactions directly on a small biopsy sample, containing 100-1000 M. leprae cells, and obtain the results within 48 hr. This means that in cases of suspected relapse, or resistance, insight can be obtained rapidly thus allowing appropriate action, such as a change of drug regimen, to be taken without delay.
Acknowledgment. We wish to thank Beth Goldstein and the Lepetit Research Center for their interest in this project. This investigation received financial support from the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases. Additional financial assistance was generously provided by the Association Française Raoul Follereau, the Institut National de la Santé et de la Recherche Médicale (CRE910604), Marion Merrell Dow, and the Institut Pasteur.
REFERENCES
1. DOCKHORN-DWORNKZAK, B., DWORNICZAK, B., BRÖMMELKAMP, L., BULLES, J., HORST, J. and BÖCKER, W. W. Non-isotopic detection of single strand conformation polymorphism (PCR-SSCP); a rapid and sensitive technique in diagnosis of phenylketonuria. Nucleic Acids Res. 19(1991)2500.
2. FRANZBLAU, S. Drug susceptibility testing of Mycobacterium leprae in the BACTEC 460 system. Anlimicrob. Agents Chemother. 33(1989)2115-2117.
3. FRANZHLAU, S. In vitro activities of aminoglycosides, lincosamides, and rifamycins against Mycobacterium leprae. Antimicrob. Agents Chemother. 35(1991)1232-1234.
4. GROSSET, J.-H., GUELPA-LAURAS, C.-C, BOHIN, P., BRUCKER, G., CARTEL, J. L., CONSTANT-DESPORTES, M., FLAGEUL, B., FRÉDÉRIC, M., GUILLAUME, J. C. and MILLAN, J. Study of 39 documented relapses of multibacillary leprosy after treatment with rifampin. Int. J. Lepr. 57(1989)607-614.
5. GROSSET, J. H. and Ji, B. Recent advances in the chemotherapy of leprosy. Lepr. Rev. 61(1990)313-329.
6. HONORÉ, N. , CHANTEAU, S., DOUCET-POPULAIRE, F., EIGLMEIER, K., GARNIER, T., GEORGES, C, LAUNOIS, P., LIMPAIHOON, P., NEWTON, S., NYANG, K., DEL PORTILLO, P., RAMESH, G. K., REDDY, T., RIEDEL, J. P., SITTISOMHUT, N., WU-HUNTER, S.and COLE, S. T. Nucleotide sequence of the first cosmid from the Mycobacterium leprae genome project: structure and function of the Rif-Str regions. Mol. Microbiol. 7(1993)207-214.
7. HONORE, N. and COLE, S. T. The molecular basis of rifampin-resistance in Mycobacterium leprae. Antimicrob. Agents Chemother. 37(1993)414-418.
8. IMHODEN, P., COLE, S., BODMER, T. and TELENTI, A. Detection of rifampin resistance in Mycobacterium tuberculosis and Mycobacterium leprae. In: Diagnostic Molecular Microbiology: Principles and Applications. Persing, D. H., Smith, T. F., Tenover, F. C. and White, T. J., eds. Washington, D.C.: American Society of Microbiology, (in press).
9. MOHAHEER, A. J., HITI, A. L. and MARTIN, W. J. Non-radiactive single strand conformation polymorphism (SSCP) using the Pharmacia "Phastsystem." Nucleic Acids Res. 19(1991)3154.
10. ORITA, M„ IWAHANA, H., KANAZAWA, H., HAYASIII, K. and SEKIYA, T. Detection of polymorphisms of human DNA by gel electrophoresis as singlestrand conformation polymorphisms. Proc. Natl. Acad. Sei USA 86 2766-2770.
11. SHEPARD, C. C. and CHANG, Y. T. Effect of several anti-leprosy drugs on multiplication of human leprosy bacilli in footpads of mice. Proc. Soc. Exp. Biol. Med. 109(1962)636-638.
12. TELENTI, A., IMBODEN, P., MARCHESI, F., LOWRIE, D., COLE, S., COLSTON, M. J., MATTER, L., SCHOPFER, K. and BODMER, T. Detection of rifampicin-rcsistance mutations in Mycobacterium tuberculosis. Lancet 1(1993)647-650.
13. WHO STUDY GROUP. Chemotherapy of leprosy for control programmes. Geneva: World Health Organization, 1982. Tech. Rep. Scr. 675.
14. WOODS, S. A. and COLE, S. T. A rapid method for the detection of potentially viable Mycobacterium leprae in human biopsies: a novel application of PCR. FEMS Microbiol. Lett. 65(1989)305-310.
1. B.Sc.; Institut Pasteur, 28 Rue du Dr. Roux, 75724 Paris 13, France.
2. Ph.D., Chef, Unité de Génétique Moléculaire Bactérienne, Institut Pasteur, 28 Rue du Dr. Roux, 75724 Paris 13, France.
3. Technical Officer; Faculté de Médecine Pitie-Salpetrière. 91 Blvd. de l'Hôpital, 75634 Paris 13, France.
4. M.D., Faculté de Médecine Pitie-Salpetrière. 91 Blvd. de l'Hôpital, 75634 Paris 13, France.
5. M.D.. Institut fur Medizinische Mikrobiologie, Universität Bern, Friedbuhlstrasse 51, Bern, Switzerland.
Reprint requests to Dr. Cole.
Received for publication on 23 March 1993.
Accepted for publication on 14 June 1993.